An Introduction to Molecular Biology/Protein synthesis
Like other biological macromolecules such as polysaccharides and nucleic acids, proteins are essential parts of living organisms and participate in virtually every process within cells. Many proteins are enzymes that catalyze biochemical reactions and are vital to metabolism. Proteins also have structural or mechanical functions, such as actin and myosin in muscle and the proteins in the cytoskeleton, which form a system of scaffolding that maintains cell shape. Other proteins are important in cell signaling, immune responses, cell adhesion, and the cell cycle. Proteins are also necessary in animals' diets, since animals cannot synthesize all the amino acids they need and must obtain essential amino acids from food. Through the process of digestion, animals break down ingested protein into free amino acids that are then used in metabolism.Proteins were first described by the Dutch chemist Gerhardus Johannes Mulder and named by the Swedish chemist Jöns Jakob Berzelius in 1838.[1]
Each protein has its own unique amino acid sequence that is specified by the nucleotide sequence of the gene encoding this protein. The genetic code is a set of three-nucleotide sets called codons and each three-nucleotide combination designates an amino acid, for example AUG (Adenine-Uracil-Guanine) is the code for methionine. Because DNA contains four nucleotides, the total number of possible codons is 64; hence, there is some redundancy in the genetic code, with some amino acids specified by more than one codon.Genes encoded in DNA are first transcribed into pre-messenger RNA (mRNA) by proteins such as RNA polymerase. Most organisms then process the pre-mRNA (also known as a primary transcript) using various forms of post-transcriptional modification to form the mature mRNA, which is then used as a template for protein synthesis by the ribosome. In prokaryotes the mRNA may either be used as soon as it is produced, or be bound by a ribosome after having moved away from the nucleoid. In contrast, eukaryotes make mRNA in the cell nucleus and then translocate it across the nuclear membrane into the cytoplasm, where protein synthesis then takes place. The rate of protein synthesis is higher in prokaryotes than eukaryotes and can reach up to 20 amino acids per second.
The mRNA is loaded onto the ribosome and is read three nucleotides at a time by matching each codon to its base pairing anticodon present on a transfer RNA (t-RNA) molecule, which carries the amino acid corresponding to the codon it recognizes. The enzyme aminoacyl tRNA synthetase "charges" the tRNA molecules with the correct amino acids. The growing polypeptide is often termed the nascent chain. Proteins are always biosynthesized from N-terminus to C-terminus.
The size of a synthesized protein can be measured by the number of amino acids it contains and by its total molecular mass, which is normally reported in units of daltons (synonymous with atomic mass units), or the derivative unit Kilodalton (kDa). Yeast proteins are on average 466 amino acids long and 53 kDa in mass. The largest known proteins are the titins, a component of the muscle sarcomere, with a molecular mass of almost 3,000 kDa and a total length of almost 27,000 amino acids.

Translation: The Synthesis of protein
[edit | edit source]The synthesis of proteins is known as translation. Translation generally occurs in the cytoplasm, where the ribosomes are located. Ribosomes are made of a small and large subunit that surround the mRNA. In translation, messenger RNA (mRNA) is decoded to produce a specific polypeptide according to the rules specified by the trinucleotide genetic code. This uses an mRNA sequence as a template to guide the synthesis of a chain of amino acids that form a protein. Translation proceeds in four phases: activation, initiation, elongation, and termination (all describing the growth of the amino acid chain, or polypeptide that is the product of translation).
In activation, the correct amino acid (AA) is joined to the correct transfer RNA (tRNA). While this is not technically a step in translation, it is required for translation to proceed. The AA is joined by its carboxyl group to the 3' OH of the tRNA by an ester bond. When the tRNA has an amino acid linked to it, it is termed "charged". Initiation involves the small subunit of the ribosome binding to 5' end of mRNA with the help of initiation factors (IF), other proteins that assist the process. Elongation occurs when the next aminoacyl-tRNA (charged tRNA) in line binds to the ribosome along with GTP and an elongation factor. Termination of the polypeptide happens when the A site of the ribosome faces a stop codon (UAA, UAG, or UGA). When this happens, no tRNA can recognize it, but releasing factor can recognize nonsense codons and causes the release of the polypeptide chain. The capacity of disabling or inhibiting translation in protein biosynthesis is used by antibiotics such as: anisomycin, cycloheximide, chloramphenicol, tetracycline, streptomycin, erythromycin, puromycin etc.[2] Each protein has its own unique amino acid sequence that is specified by the nucleotide sequence of the gene encoding this protein. The genetic code is a set of three-nucleotide sets called codons and each three-nucleotide combination designates an amino acid, for example AUG (Adenine-Uracil-Guanine) is the code for methionine. Because DNA contains four nucleotides, the total number of possible codons is 64; hence, there is some redundancy in the genetic code, with some amino acids specified by more than one codon. Genes encoded in DNA are first transcribed into pre-messenger RNA (mRNA) by proteins such as RNA polymerase. Most organisms then process the pre-mRNA (also known as a primary transcript) using various forms of post-transcriptional modification to form the mature mRNA, which is then used as a template for protein synthesis by the ribosome. In prokaryotes the mRNA may either be used as soon as it is produced, or be bound by a ribosome after having moved away from the nucleoid. In contrast, eukaryotes make mRNA in the cell nucleus and then translocate it across the nuclear membrane into the cytoplasm, where protein synthesis then takes place. The rate of protein synthesis is higher in prokaryotes than eukaryotes and can reach up to 20 amino acids per second. We should always remember that following antibiotics inhibit the protein synthesis e.g. anisomycin, cycloheximide, chloramphenicol, tetracycline, streptomycin, erythromycin, puromycin etc.
The mRNA is loaded onto the ribosome and is read three nucleotides at a time by matching each codon to its base pairing anticodon present on a transfer RNA (t-RNA) molecule, which carries the amino acid corresponding to the codon it recognizes. The enzyme aminoacyl tRNA synthetase "charges" the tRNA molecules with the correct amino acids. The growing polypeptide is often termed the nascent chain. Proteins are always biosynthesized from N-terminus to C-terminus. The size of a synthesized protein can be measured by the number of amino acids it contains and by its total molecular mass, which is normally reported in units of daltons (synonymous with atomic mass units), or the derivative unit Kilodalton (kDa). Yeast proteins are on average 466 amino acids long and 53 kDa in mass. The largest known proteins are the titins, a component of the muscle sarcomere, with a molecular mass of almost 3,000 kDa and a total length of almost 27,000 amino acids.[3]
tRNA (Transfer RNA)
[edit | edit source]
tRNA appears like cloverleaf structure. Its anticodon arm is a 5 base pair (bp) stem whose loop contains the anticodon while its D arm is a 4 bp stem ending in a loop that often contains dihydrouridine. T arm of tRNA is a 5 bp stem containing the sequence TΨC where Ψ is a pseudouridine.In eukaryotic cells, tRNAs are transcribed by RNA pol III as pre-tRNAs in the nucleus. tRNA is a small RNA molecule usually 73-95 nucleotides long. RNA polymerase III recognizes two internal promoter sequences (A-box B internal promoter) inside tRNA genes. The first promoter begins at nucleotide 8 of mature tRNAs and the second promoter is located 30-60 nucleotides downstream of the first promoter. The transcription terminates after a stretch of four or more thymidines.
Pre-tRNAs undergo extensive modifications inside the nucleus. Some pre-tRNAs contain introns; in bacteria these self-splice, whereas in eukaryotes and archaea they are removed by tRNA splicing endonuclease.The 5' sequence is removed by RNase P, whereas the 3' end is removed by the tRNase Z enzyme. A notable exception is in the archaeon Nanoarchaeum equitans which does not possess an RNase P enzyme and has a promoter placed such that transcription starts at the 5' end of the mature tRNA.. The non-templated 3' CCA tail is added by a nucleotidyl transferase. Before tRNAs are exported into the cytoplasm by Los1/Xpo-t, tRNAs are aminoacylated. The order of the processing events is not conserved. For example in yeast, the splicing is not carried out in the nucleus but at the cytoplasmic side of mitochondrial membranes.[4]
Ribosome
[edit | edit source]Ribosomes are the components of cells that make proteins from all amino acids. One of the central tenets of biology, often referred to as the "central dogma," is that DNA is used to make RNA, which, in turn, is used to make protein. The DNA sequence in genes is copied into a messenger RNA (mRNA). Ribosomes then read the information in this RNA and use it to create proteins. This process is known as translation; i.e., the ribosome "translates" the genetic information from RNA into proteins. Ribosomes do this by binding to an mRNA and using it as a template for the correct sequence of amino acids in a particular protein. The amino acids are attached to transfer RNA (tRNA) molecules, which enter one part of the ribosome and bind to the messenger RNA sequence. The attached amino acids are then joined together by another part of the ribosome. The ribosome moves along the mRNA, "reading" its sequence and producing a chain of amino acids.
Ribosomes are made from complexes of RNAs and proteins. Ribosomes are divided into two subunits, one larger than the other. The smaller subunit binds to the mRNA, while the larger subunit binds to the tRNA and the amino acids. When a ribosome finishes reading a mRNA, these two subunits split apart. Ribosomes have been classified as ribozymes, since the ribosomal RNA seems to be most important for the peptidyl transferase activity that links amino acids together.[5]
Ribosomes from bacteria, archaea and eukaryotes (the three domains of life on Earth), have significantly different structures and RNA sequences. These differences in structure allow some antibiotics to kill bacteria by inhibiting their ribosomes, while leaving human ribosomes unaffected. The ribosomes in the mitochondria of eukaryotic cells resemble those in bacteria, reflecting the likely evolutionary origin of this organelle. The word ribosome comes from ribonucleic acid and the Greek: soma (meaning body).
Structure of the Ribosome
[edit | edit source]
A svedberg (symbol S, sometimes Sv, not to be confused with Sv for the SI unit sievert as well as the non-SI sverdrup) is a non-SI physical unit used for sedimentation coefficients. It characterizes the behaviour of a particle type in sedimentation processes, notably centrifugation. The svedberg is technically a measure of time, and is defined as exactly 10-13 seconds (100 fs).
The unit is named after the Swedish chemist Theodor Svedberg (1884-1971), winner of the Nobel prize in chemistry in 1926 for his work in the chemistry of colloids and his invention of the ultracentrifuge. Bigger particles tend to sediment faster and thus have higher svedberg values. Sedimentation coefficients are, however, not additive. Sedimentation rate does not depend only on the mass or volume of a particle, and when two particles bind together there is inevitably a loss of surface area. Thus when measured separately they will have svedberg values that may not add up to that of the bound particle. The svedberg is the most important measure used to distinguish ribosomes, which are important in phylogenetic studies.
The ribosomal subunits of prokaryotes and eukaryotes are quite similar.The unit of measurement is the Svedberg unit, a measure of the rate of sedimentation in centrifugation rather than size and accounts for why fragment names do not add up (70S is made of 50S and 30S). Prokaryotes have 70S ribosomes, each consisting of a small (30S) and a large (50S) subunit. Their large subunit is composed of a 5S RNA subunit (consisting of 120 nucleotides), a 23S RNA subunit (2900 nucleotides) and 34 proteins. The 30S subunit has a 1540 nucleotide RNA subunit (16S) bound to 21 proteins. Eukaryotes have 80S ribosomes, each consisting of a small (40S) and large (60S) subunit. Their large subunit is composed of a 5S RNA (120 nucleotides), a 28S RNA (4700 nucleotides), a 5.8S subunit (160 nucleotides) and ~49 proteins. The 40S subunit has a 1900 nucleotide (18S) RNA and ~33 proteins.
The ribosomes found in chloroplasts and mitochondria of eukaryotes also consist of large and small subunits bound together with proteins into one 70S particle. These organelles are believed to be descendants of bacteria (see Endosymbiotic theory) and as such their ribosomes are similar to those of bacteria. The various ribosomes share a core structure, which is quite similar despite the large differences in size. Much of the RNA is highly organized into various tertiary structural motifs, for example pseudoknots that exhibit coaxial stacking. The extra RNA in the larger ribosomes is in several long continuous insertions, such that they form loops out of the core structure without disrupting or changing it. All of the catalytic activity of the ribosome is carried out by the RNA; the proteins reside on the surface and seem to stabilize the structure.
The differences between the bacterial and eukaryotic ribosomes are exploited by pharmaceutical chemists to create antibiotics that can destroy a bacterial infection without harming the cells of the infected person. Due to the differences in their structures, the bacterial 70S ribosomes are vulnerable to these antibiotics while the eukaryotic 80S ribosomes are not. Even though mitochondria possess ribosomes similar to the bacterial ones, mitochondria are not affected by these antibiotics because they are surrounded by a double membrane that does not easily admit these antibiotics into the organelle.

Ribogenesis in Prokaryotes There are 52 genes that encode the ribosomal proteins and they can be found in 20 operons within prokaryotic DNA. Regulation of ribosome synthesis hinges on the regulation of the rRNA itself.First, a reduction in aminoacyl-tRNA will cause the prokaryotic cell to respond by lowering transcription and translation. This occurs through a series of steps, beginning with stringent factor binding to ribosomes and catalyzing the reaction:
GTP + ATP --> pppGpp + AMP
The γ-phosphate is then removed and ppGpp will bind to and inhibit RNA polymerase. This binding causes a reduction in rRNA transcription. A reduced amount of rRNA means that ribosomal proteins (r-proteins) will be translated but will not have an rRNA to bind to. Instead, they will negatively feedback and bind to their own mRNA, repressing r-protein synthesis. Note that r-proteins preferentially bind to its complementary rRNA if it is present, rather than mRNA. The ribosome operons also include the genes for RNA polymerase and elongation factors (used in RNA translation). Regulation of all of these genes at once illustrate the coupling between transcription and translation in prokaryotes.
Ribogenesis in Eukaryotes Ribosomal protein synthesis in eukaryotes occurs, like most protein synthesis, in the cytoplasm just outside the nucleus. Individual large and small units are synthesized and imported into the nucleus through nuclear pores. These pores have a diameter of 120 nm and import 560,000 ribosomal proteins per minute into the nucleus with active transport. See nuclear import for more about the movement of the ribosomal proteins into the nucleus. Ribosomal RNA (rRNA) is transcribed at the nucleolus, at a high speed, which contains all 45S rRNA genes. After transcription, the rRNA is put together with the ribosomal subunits to make a functioning ribosome.
Kozak consensus sequence
[edit | edit source]Kozak consensus sequence on an mRNA molecule is recognized by the ribosome as the translational start site, from which a protein is coded by that mRNA molecule. The ribosome requires this sequence, or a possible variation to initiate translation. The Kozak sequence is not to be confused with the ribosomal binding site (RBS), that being either the 5' cap of a messenger RNA or an Internal Ribosome Entry Site (IRES). In vivo, this site is often not matched exactly on different mRNAs and the amount of protein synthesized from a given mRNA is dependent on the strength of the Kozak sequence. Some nucleotides in this sequence are more important than others: the AUG is most important because it is the actual initiation codon encoding a methionine amino acid at the N-terminus of the protein. (Rarely, CTG is used as an initiation codon, encoding a leucine instead of its typical methionine.) The A nucleotide of the "AUG" is referred to as number 1. For a 'strong' consensus, the nucleotides at positions +4 (i.e. G in the consensus) and -3 (i.e. either A or G in the consensus) relative to the number 1 nucleotide must both match the consensus (there is no number 0 position). An 'adequate' consensus has only 1 of these sites, while a 'weak' consensus has neither. The cc at -1 and -2 are not as conserved, but contribute to the overall strength. There is also evidence that a G in the -6 position is important in the initiation of translation.
There are examples in vivo of each of these types of Kozak consensus, and they probably evolved as yet another mechanism of gene regulation. Lmx1b is an example of a gene with a weak Kozak consensus sequence. For initiation of translation from such a site, other features are required in the mRNA sequence in order for the ribosome to recognize the initiation codon.[6]
Synthesis of protein in Prokaryotes
[edit | edit source]Initiation
[edit | edit source]
Initiation of translation in prokaryotes involves the assembly of the components of the translation system which are: the two ribosomal subunits (50S & 30S subunits), the mRNA to be translated, the first (formyl) aminoacyl tRNA (the tRNA charged with the first amino acid), GTP (as a source of energy), and three initiation factors (Prokaryotic initiation factor-1 or IF1, Prokaryotic initiation factor-2 or IF2, and Prokaryotic initiation factor-3 or IF3 which help the assembly of the initiation complex.[7]
The ribosome has three active site|sites: the A site, the P site, and the E site. The A site is the point of entry for the aminoacyl tRNA (except for the first aminoacyl tRNA, fMet-tRNAfMet, which enters at the P site). The P site is where the peptidyl tRNA is formed in the ribosome. And the E site which is the exit site of the now uncharged tRNA after it gives its amino acid to the growing peptide chain.
Elongation
[edit | edit source]Elongation of the polypeptide chain involves addition of amino acids to the carboxyl end of the growing chain. The growing protein exits the ribosome through the polypeptide exit tunnel in the large subunit.[8] In prokaryotes, three elongation factors are required for translation: EF-Tu, EF-Ts, and EF-G.
EF-Tu (elongation factor thermo unstable) mediates the entry of the aminoacyl tRNA into a free site of the ribosome.
EF-Ts serves as the guanine nucleotide exchange factor for EF-Tu, catalyzing the release of GDP from EF-Tu.
EF-G catalyzes the translocation of the tRNA and mRNA down the ribosome at the end of each round of polypeptide elongation.
Elongation starts when the fmet-tRNA enters the P site, causing a conformational change which opens the A site for the new aminoacyl-tRNA to bind. This binding is facilitated by elongation factor-Tu (EF-Tu), a small GTPase. Now the P site contains the beginning of the peptide chain of the protein to be encoded and the A site has the next amino acid to be added to the peptide chain. The growing polypeptide connected to the tRNA in the P site is detached from the tRNA in the P site and a peptide bond is formed between the last amino acids of the polypeptide and the amino acid still attached to the tRNA in the A site. This process, known as peptide bond formation, is catalyzed by a ribozyme (the 23S ribosomal RNA in the 50S ribosomal subunit). Now, the A site has the newly formed peptide, while the P site has an uncharged tRNA (tRNA with no amino acids). In the final stage of elongation, translocation, the ribosome moves 3 nucleotides towards the 3'end of mRNA. Since tRNAs are linked to mRNA by codon-anticodon base-pairing, tRNAs move relative to the ribosome taking the nascent polypeptide from the A site to the P site and moving the uncharged tRNA to the E exit site. This process is catalyzed by elongation factor G (EF-G).
The ribosome continues to translate the remaining codons on the mRNA as more aminoacyl-tRNA bind to the A site, until the ribosome reaches a stop codon on mRNA(UAA, UGA, or UAG).[9]
EF-Tu
EF-Tu (elongation factor thermo unstable) is one of the prokaryotic elongation factors. The prokaryotic factor EF-Tu mediates the entry of the aminoacyl tRNA into a free site of the ribosome. EF-Tu functions by binding an aminoacylated, or charged, tRNA molecule in the cytoplasm. This complex transiently enters the ribosome, with the tRNA anticodon domain associating with the mRNA codon in the ribosomal A site. If the codon-anticodon pairing is correct, EF-Tu hydrolyzes guanosine triphosphate (GTP) into guanosine diphosphate (GDP) and inorganic phosphate, and changes in conformation to dissociate from the tRNA molecule. The aminoacyl tRNA then fully enters the A site, where its amino acid is brought near the P-site polypeptide and the ribosome catalyzes the covalent transfer of the amino acid onto the polypeptide. EF-Tu contributes to translational accuracy in three ways. It delays GTP hydrolysis if the tRNA in the ribosome’s A site does not match the mRNA codon, thus preferentially increasing the likelihood for the incorrect tRNA to leave the ribosome. It also adds a second delay (regardless of tRNA matching) after freeing itself from tRNA, before the aminoacyl tRNA fully enters the A site. This delay period is a second opportunity for incorrectly-paired tRNA (and their bound amino acids) to move out of the A site before the incorrect amino acid is irreversibly added to the polypeptide chain. A third mechanism is the less well understood function of EF-Tu to crudely check amino acid-tRNA associations, and reject complexes where the amino acid is not bound to the correct tRNA coding for it.
EF-Ts
EF-Ts (elongation factor thermo stable) is one of the prokaryotic elongation factors. EF-Ts serves as the guanine nucleotide exchange factor for EF-Tu (elongation factor thermo unstable), catalyzing the release of guanosine diphosphate from EF-Tu. This enables EF-Tu to bind to a new guanosine triphosphate molecule, release EF-Ts, and go on to catalyze another aminoacyl tRNA addition.
EF-G
The factor EF-G catalyzes the translocation of the tRNA and mRNA down the ribosome at the end of each round of polypeptide elongation. Homologous to EF-Tu + tRNA, EF-G also binds to the ribosome in its GTP-bound state. When it associates with the A site, EF-G causes the tRNA previously occupying that site to occupy an intermediate A/P position (bound to the A site of the small ribosomal subunit and to the P site of the large subunit), and the tRNA in the P site is shifted to a P/E hybrid state. EF-G hydrolysis of GTP causes a conformation change that forces the A/P tRNA to fully occupy the P site, the P/E tRNA to fully occupy the E site (and exit the ribosome complex), and the mRNA to shift three nucleotides down relative to the ribosome due to its association with these tRNA molecules. The GDP-bound EF-G molecule then dissociates from the complex, leaving another free A-site where the elongation cycle can start again. Apart from its role in translocation, EF-G, working together with Ribosome Recycling Factor promotes ribosome recycling in a GTP-dependent manner

Termination
[edit | edit source]Termination occurs when one of the three termination codons. moves into the A site. These codons are not recognized by any tRNAs. Instead, they are recognized by proteins called release factors, namely RF1 (recognizing the UAA and UAG stop codons) or RF2 (recognizing the UAA and UGA stop codons). These factors trigger the hydrolysis of the ester bond in peptidyl-tRNA and the release of the newly synthesized protein from the ribosome. A third release factor RF-3 catalyzes the release of RF-1 and RF-2 at the end of the termination process.
Recycling
[edit | edit source]The post-termination complex formed by the end of the termination step consists of mRNA with the termination codon at the A-site, an uncharged tRNA in the P site, and the intact 70S ribosome. Ribosome recycling step is responsible for the disassembly of the post-termination ribosomal complex.[10] Once the nascent protein is released in termination, Ribosome Recycling Factor and Elongation Factor G (EF-G) function to release mRNA and tRNAs from ribosomes and dissociate the 70S ribosome into the 30S and 50S subunits. IF3 then replaces the deacylated tRNA releasing the mRNA. All translational components are now free for additional rounds of translation.
Polysomes
[edit | edit source]Translation is carried out by more than one ribosome simultaneously. Because of the relatively large size of ribosomes, they can only attach to sites on mRNA 35 nucleotides apart. The complex of one mRNA and a number of ribosomes is called a polysome or polyribosome.
Synthesis of protein in Eukaryotes
[edit | edit source]
Initiation
[edit | edit source]Eukaryotic initiation factors (eIF) are proteins involved in the initiation phase of eukaryotic translation. They function in forming a complex with the 40S ribosomal subunit and Met-tRNAi called the 43S preinitation complex (PIC), recognizing the 5' cap structure of mRNA and recruiting the 43S PIC to mRNA, promoting ribosomal scanning of mRNA and regulating recognition of the AUG initiation codon, and joining of the 60S ribosomal subunit to create the 80S ribosome. There exist many more eukaryotic initiation factors than prokaryotic initiation factors due to greater biological complexity of eukaryotic cells. The protein RLI is known to have an essential, probably catalytic role in the formation of initiation complexes as well.
eIF4 (eIF4F)
The eIF4 initiation factors include eIF4A, eIF4B, eIF4E, and eIF4G. eIF4F is often used to refer to the complex of eIF4A, eIF4E, and eIF4G. eIF4G is a scaffolding protein that interacts with eIF3 (see below), as well as the other members of the eIF4F complex. eIF4E recognizes and binds to the 5' cap structure of mRNA, while eIF4G binds to Poly(A)-binding protein which binds the poly(A) tail, circularizing and activating the bound mRNA. eIF4A – a DEAD box RNA helicase – is important for resolving mRNA secondary structures. eIF4B contains two RNA-binding domains – one non-specifically interacts with mRNA, whereas the second specifically binds the 18S portion of the small ribosomal subunit. It acts as an anchor, as well as a critical co-factor for eIF4A. It is a substrate of S6K, and, when phosphorylated, it promotes the formation of the pre-initiation complex. In vertebrates, eIF4H is an additional initiation factor with similar function to eIF4B.[11]
eIF1 & eIF3
eIF1, eIF1A, and eIF3, all bind to the ribosome subunit-mRNA complex. They have been implicated in preventing the large ribosomal subunit from binding the small subunit before it is ready to commence elongation. In mammals, eIF3 is the largest scaffolding initiation factor, made up of 13 subunits (a-m). It is roughly ~750 kDa and it controls the assembly of 40S ribosomal subunit on mRNA that have a 5' cap or an IRES. eIF3 uses the eIF4F complex or IRES (Internal Ribosomal Entry Site) from viruses to position the mRNA strand near the exit site of the 40S ribosome subunit, thus promoting the assembly of the pre-initiation complex. In many cancers, eIF3 is overexpressed. Under serum-deprived conditions (inactive state), eIF3 is bound to S6K1. On stimulation either by mitogens, growth factors, or drugs, mTOR/Raptor complex gets activated and, in turn, binds and phosphorylates S6K1 on T389 (linker region), causing a conformational change that causes the kinase S6K1 to dissociate from eIF3. The T389 phosphorylated S6k1 is then further phosphorylated by PDK1 on T229. This second phosphorylation fully activates the S6K1 kinase, which can then phosphorylate eIF4B, S6 and other protein targets.[12]
eIF2
eIF2 is a GTP-binding protein responsible for bringing the initiator tRNA to the P-site of the pre-initiation complex. It has specificity for the methionine-charged initiator tRNA, which is distinct from other methionine-charged tRNAs specific for elongation of the polypeptide chain. Once it has placed the initiator tRNA on the AUG start codon in the P-site, it hydrolyzes GTP into GDP, and dissociates. This hydrolysis, also signals for the dissociation of eIF3, eIF1, and eIF1A, and allows the large subunit to bind. This signals the beginning of elongation. eIF2 has three subunits, eIF2-α, β, and γ. The former is of particular importance for cells that may need to turn off protein synthesis globally. When phosphorylated, it sequesters eIF2B (not to be confused with beta), a GEF. Without this GEF, GDP cannot be exchanged for GTP, and translation is repressed. eIF2α-induced translation repression occurs in reticulocytes when starved for iron. In addition, protein kinase R (PKR) phosphorylates eIF2α when dsRNA is detected in many multicellular organisms, leading to cell death.[13]
eIF5 & eIF5B
eIF5A is a GTPase-activating protein, which helps the large ribosomal subunit associate with the small subunit. It is required for GTP-hydrolysis by eIF2 and contains the unusual amino acid hypusine. eIF5B is a GTPase, and is involved in assembly of the full ribosome (which requires GTP hydrolysis).
eIF6 eIF6 performs the same inhibition of ribosome assembly as eIF3, but binds with the large subunit.
The process of initiation of translation in eukaryotes depend up on mRNA capping.
The cap-independent initiation
[edit | edit source]The best studied example of the cap-independent mode of translation initiation in eukaryotes is the Internal Ribosome Entry Site (IRES) approach. What differentiates cap-independent translation from cap-dependent translation is that cap-independent translation does not require the ribosome to start scanning from the 5' end of the mRNA cap until the start codon. The ribosome can be trafficked to the start site by ITAFs (IRES trans-acting factors) bypassing the need to scan from the 5' end of the untranslated region of the mRNA. This method of translation has been recently discovered, and has found to be important in conditions that require the translation of specific mRNAs, despite cellular stress or the inability to translate most mRNAs. Examples include factors responding to apoptosis, stress-induced responses.
Cap-dependent initiation
[edit | edit source]Initiation of translation usually involves the interaction of certain key proteins with a special tag bound to the 5'-end of an mRNA molecule, the 5' cap. The protein factors bind the small ribosomal subunit (also referred to as the 40S subunit), and these initiation factors hold the mRNA in place. The eukaryotic Initiation Factor 3 (eIF3) is associated with the small ribosomal subunit, and plays a role in keeping the large ribosomal subunit from prematurely binding. eIF3 also interacts with the eIF4F complex which consists of three other initiation factors: eIF4A, eIF4E and eIF4G. eIF4G is a scaffolding protein which directly associates with both eIF3 and the other two components. eIF4E is the cap-binding protein. It is the rate-limiting step of cap-dependent initiation, and is often cleaved from the complex by some viral proteases to limit the cell's ability to translate its own transcripts. This is a method of hijacking the host machinery in favor of the viral (cap-independent) messages. eIF4A is an ATP-dependent RNA helicase, which aids the ribosome in resolving certain secondary structures formed by the mRNA transcript. There is another protein associated with the eIF4F complex called the Poly(A)-binding protein (PABP), which binds the poly-A tail of most eukaryotic mRNA molecules. This protein has been implicated in playing a role in circularization of the mRNA during translation. This pre-initiation complex (43S subunit, or the 40S and mRNA) accompanied by the protein factors move along the mRNA chain towards its 3'-end, scanning for the 'start' codon (typically AUG) on the mRNA, which indicates where the mRNA will begin coding for the protein. In eukaryotes and archaea, the amino acid encoded by the start codon is methionine. The initiator tRNA charged with Met forms part of the ribosomal complex and thus all proteins start with this amino acid (unless it is cleaved away by a protease in subsequent modifications). The Met-charged initiator tRNA is brought to the P-site of the small ribosomal subunit by eukaryotic Initiation Factor 2 (eIF2). It hydrolyzes GTP, and signals for the dissociation of several factors from the small ribosomal subunit which results in the association of the large subunit (or the 60S subunit). The complete ribosome (80S) then commences translation elongation, during which the sequence between the 'start' and 'stop' codons is translated from mRNA into an amino acid sequence—thus a protein is synthesized.
Regulation of protein synthesis is dependent on phosphorylation of initiation factor eIF2 which is a part of the met-tRNAi complex. When large numbers of eIF2 are phosphorylated, protein synthesis is inhibited. This would occur if there is amino acid starvation or there has been a virus infection. However naturally a small percentage is of this initiation factor is phosphorylated. Another regulator is 4EBP which binds to the initiation factor eIF4E found on the 5’ cap on mRNA stopping protein synthesis. To oppose the effects of the 4EBP growth factors phosphorylate 4EBP reducing its affinity for eIF4E and permitting protein synthesis.[14]
Elongation
[edit | edit source]Eukaryotic elongation factors are very similar to those in prokaryotes. Elongation in eukaryotes is carried out with two elongation factors: eEF-1 and eEF-2. The first is eEF-1, and has two subunits, α and βγ. α acts as counterpart to prokaryotic EF-Tu, mediating the entry of the aminoacyl tRNA into a free site of the ribosome. βγ acts as counterpart to prokaryotic EF-Ts, serving as the guanine nucleotide exchange factor for α, catalyzing the release of GDP from α. The second elongation factor is eEF-2, the counterpart to prokaryotic EF-G, catalyzing the translocation of the tRNA and mRNA down the ribosome at the end of each round of polypeptide elongation.
At the end of the initiation step, the mRNA is positioned so that the next codon can be translated during the elongation stage of protein synthesis. The initiator tRNA occupies the P site in the ribosome, and the A site is ready to receive an aminoacyl-tRNA. During chain elongation, each additional amino acid is added to the nascent polypeptide chain in a three-step microcycle. The steps in this microcycle are (1) positioning the correct aminoacyl-tRNA in the A site of the ribosome, (2) forming the peptide bond and (3) shifting the mRNA by one codon relative to the ribosome. The translation machinery works relatively slowly compared to the enzyme systems that catalyze DNA replication. Proteins in prokaryotes are synthesized at a rate of only 18 amino acid residues per second, whereas bacterial replisomes synthesize DNA at a rate of 1000 nucleotides per second. This difference in rate reflects, in part, the difference between polymerizing four types of nucleotides to make nucleic acids and polymerizing 20 types of amino acids to make proteins. Testing and rejecting incorrect aminoacyl-tRNA molecules takes time and slows protein synthesis. The rate of transcription in prokaryotes is approximately 55 nucleotides per second, which corresponds to about 18 codons per second, or the same rate at which the mRNA is translated. In bacteria, translation initiation occurs as soon as the 5' end of an mRNA is synthesized, and translation and transcription are coupled. This tight coupling is not possible in eukaryotes because transcription and translation are carried out in separate compartments of the cell (the nucleus and cytoplasm). Eukaryotic mRNA precursors must be processed in the nucleus (e.g. capping, polyadenylation, splicing) before they are exported to the cytoplasm for translation.[15]
Termination
[edit | edit source]Termination of elongation is dependent on eukaryotic release factors. The process of termination is similar to that of prokaryotic termination.
Structure of protein
[edit | edit source]
Primary structure of protein
[edit | edit source]The primary structure refers to the sequence of the different amino acids of the peptide or protein. The primary structure is held together by covalent or peptide bonds, which are made during the process of protein biosynthesis or translation. The two ends of the polypeptide chain are referred to as the carboxyl terminus (C-terminus) and the amino terminus (N-terminus) based on the nature of the free group on each extremity. Counting of residues always starts at the N-terminal end (NH2-group), which is the end where the amino group is not involved in a peptide bond. The primary structure of a protein is determined by the gene corresponding to the protein.
A specific sequence of nucleotides in DNA is transcribed into mRNA, which is read by the ribosome in a process called translation. The sequence of a protein is unique to that protein, and defines the structure and function of the protein. The sequence of a protein can be determined by methods such as Edman degradation or tandem mass spectrometry. Often however, it is read directly from the sequence of the gene using the genetic code. Post-translational modifications such as disulfide formation, phosphorylations and glycosylations are usually also considered a part of the primary structure, and cannot be read from the gene.[16]
Secondary structure of protein
[edit | edit source]In molecular biology and structural biology, secondary structure is the general three-dimensional form of local segments of biopolymers such as proteins and nucleic acids (DNA/RNA). It does not, however, describe specific atomic positions in three-dimensional space, which are considered to be tertiary structure.
Secondary structure can be formally defined by the hydrogen bonds of the biopolymer, as observed in an atomic-resolution structure. In proteins, the secondary structure is defined by the patterns of hydrogen bonds between backbone amide and carboxyl groups. In nucleic acids, the secondary structure is defined by the hydrogen bonding between the nitrogenous bases. The hydrogen bonding patterns may be significantly distorted, which makes an automatic determination of secondary structure difficult. The secondary structure may be also defined based on the regular pattern of backbone dihedral angles in a particular region of the Ramachandran plot; thus, a segment of residues with such dihedral angles may be called a helix, regardless of whether it has the correct hydrogen bonds. The secondary structure may be also provided by crystallographers in the corresponding PDB file.
The rough secondary-structure content of a biopolymer (e.g., "this protein is 40% α-helix and 20% β-sheet.") can often be estimated spectroscopically. For proteins, a common method is far-ultraviolet (far-UV, 170-250 nm) circular dichroism. A pronounced double minimum at 208 and 222 nm indicate α-helical structure, whereas a single minimum at 204 nm or 217 nm reflects random-coil or β-sheet structure, respectively. A less common method is infrared spectroscopy, which detects differences in the bond oscillations of amide groups due to hydrogen-bonding. Finally, secondary-structure contents may be estimated accurately using the chemical shifts of an unassigned NMR spectrum. Secondary structure was introduced by Kaj Ulrik Linderstrøm-Lang at Stanford in 1952.[17]
| Geometry attribute | α-helix | 310 helix | π-helix |
|---|---|---|---|
| Residues per turn | 3.6 | 3.0 | 4.4 |
| Translation per residue | 1.5Å | 2.0|Å | 1.1 Å |
| Radius of helix | 2.3 Å | 1.9 Å | 2.8 Å |
| Pitch | 5.4 Å | 6.0 Å | 4.8 Å |
Tertiary structure of protein
[edit | edit source]Tertiary structure refers to three-dimensional structure of a single protein molecule. The alpha-helices and beta-sheets are folded into a compact globule. The folding is driven by the non-specific hydrophobic interactions (the burial of hydrophobic residues from water), but the structure is stable only when the parts of a protein domain are locked into place by specific tertiary interactions, such as salt bridges, hydrogen bonds, and the tight packing of side chains and disulfide bonds. The disulfide bonds are extremely rare in cytosolic proteins, since the cytosol is generally a reducing environment.
Quaternary structure of protein
[edit | edit source]Quaternary structure is a larger assembly of several protein molecules or polypeptide chains, usually called subunits in this context. The quaternary structure is stabilized by the same non-covalent interactions and disulfide bonds as the tertiary structure. Complexes of two or more polypeptides (i.e. multiple subunits) are called multimers. Specifically it would be called a dimer if it contains two subunits, a trimer if it contains three subunits, and a tetramer if it contains four subunits. The subunits are frequently related to one another by symmetry operations, such as a 2-fold axis in a dimer. Multimers made up of identical subunits are referred to with a prefix of "homo-" (e.g. a homotetramer) and those made up of different subunits are referred to with a prefix of "hetero-" (e.g. a heterotetramer, such as the two alpha and two beta chains of hemoglobin). Many proteins do not have the quaternary structure and function as monomers.
Drugs which inhibit protein synthesis
[edit | edit source]In general, protein synthesis inhibitors work at different stages of prokaryotic mRNA translation into proteins, like initiation, elongation (including aminoacyl tRNA entry, proofreading, peptidyl transfer, and ribosomal translocation) and termination:
Earlier stages
Rifampicin inhibits prokaryotic DNA transcription into mRNA by inhibiting DNA-dependent RNA polymerase by binding its beta-subunit.
Initiation Linezolid acts at the initiation stage, probably by preventing the formation of the initiation complex, although the mechanism is not fully understood.
Aminoacyl tRNA entry
Tetracyclines and Tigecycline(a glycylcycline related to tetracyclines) block the A site on the ribosome, preventing the binding of aminoacyl tRNAs.
Proofreading
Aminoglycosides, among other potential mechanisms of action, interfere with the proofreading process, causing increased rate of error in synthesis with premature termination.
Peptidyl transfer
Chloramphenicol blocks the peptidyl transfer step of elongation on the 50S ribosomal subunit in both bacteria and mitochondria. Macrolides (as well as inhibiting ribosomal translocation and other potential mechanisms) bind to the 50s ribosomal subunits, inhibiting peptidyl transfer. Quinupristin/dalfopristin act synergistically, with dalfopristin, enhancing the binding of quinupristin, as well as inhibiting peptidyl transfer. Quinupristin binds to binds to a nearby site on the 50S ribosomal subunit and prevents elongation of the polypeptide, as well as causing incomplete chains to be released.
Ribosomal translocation
Clindamycin, among other potential mechanisms. Aminoglycosides and macrolides among other potential mechanisms of action, have evidence of inhibition of ribosomal translocation. Fusidic acid prevents the turnover of elongation factor G (EF-G) from the ribosome.
Termination
Puromycin has a structure similar to that of the tyrosinyl aminoacyl-tRNA. Thus, it binds to the ribosomal A site and participates in peptide bond formation, producing peptidyl-puromycin. However, it does not engage in translocation and quickly dissociates from the ribosome, causing a premature termination of polypeptide synthesis. Macrolides and clindamycin (both also having other potential mechanisms) cause premature dissociation of the peptidyl-tRNA from the ribosome. Streptogramins also cause premature release of the peptide chain.
Protein synthesis inhibitors of unspecified mechanism
Retapamulin
Binding site
The following antibiotics bind to the 30S subunit of the ribosome:
Aminoglycosides
Tetracyclines
The following antibiotics bind to the 50S ribosomal subunit:
Chloramphenicol
Erythromycin
Clindamycin
Linezolid
Telithromycin
Streptogramins
Retapamulin
Posttranslational modification of protein
[edit | edit source]Posttranslational modification (PTM) is the chemical modification of a protein after its translation. It is one of the later steps in protein biosynthesis for many proteins. PTMs involving addition of functional groups
PTMs involving addition by an enzyme in vivo acylation, e.g. O-acylation (esters), N-acylation (amides), S-acylation (thioesters) acetylation, the addition of an acetyl group, either at the N-terminus of the protein or at lysine residues. See also histone acetylation. The reverse is called deacetylation. formylation lipoylation, attachment of a lipoate (C8) functional group myristoylation, attachment of myristate, a C14 saturated acid palmitoylation, attachment of palmitate, a C16 saturated acid alkylation, the addition of an alkyl group, e.g. methyl, ethyl methylation the addition of a methyl group, usually at lysine or arginine residues. The reverse is called demethylation. isoprenylation or prenylation, the addition of an isoprenoid group (e.g. farnesol and geranylgeraniol) farnesylation geranylgeranylation amidation at C-terminus amino acid addition arginylation, a tRNA-mediation addition polyglutamylation, covalent linkage of glutamic acid residues to tubulin and some other proteins. (See tubulin polyglutamylase) polyglycylation, covalent linkage of one to more than 40 glycine residues to the tubulin C-terminal tail diphthamide formation gamma-carboxylation dependent on Vitamin K glycosylation, the addition of a glycosyl group to either asparagine, hydroxylysine, serine, or threonine, resulting in a glycoprotein. Distinct from glycation, which is regarded as a nonenzymatic attachment of sugars. polysialylation, addition of polysialic acid, PSA, to NCAM glypiation, glycosylphosphatidylinositol (GPI) anchor formation heme moiety may be covalently attached hydroxylation hypusine formation (on conserved lysine of [EIF5A] and aIF5a) iodination (e.g. of thyroid hormones) nucleotides or derivatives thereof may be covalently attached adenylation ADP-ribosylation flavin attachment nitrosylation S-glutathionylation oxidation phosphopantetheinylation, the addition of a 4'-phosphopantetheinyl moiety from coenzyme A, as in fatty acid, polyketide, non-ribosomal peptide and leucine biosynthesis phosphorylation, the addition of a phosphate group, usually to serine, tyrosine, threonine or histidine pyroglutamate formation sulfation, the addition of a sulfate group to a tyrosine. selenoylation (co-translational incorporation of selenium in selenoproteins) PTMs involving non-enzymatic additions in vivo glycation, the addition of a sugar molecule to a protein without the controlling action of an enzyme. PTMs involving non-enzymatic additions in vitro biotinylation, acylation of conserved lysine residues with a biotin appendage pegylation PTMs involving addition of other proteins or peptides
ISGylation, the covalent linkage to the ISG15 protein (Interferon-Stimulated Gene 15) SUMOylation, the covalent linkage to the SUMO protein (Small Ubiquitin-related MOdifier) ubiquitination, the covalent linkage to the protein ubiquitin. Neddylation, the covalent linkage to Nedd PTMs involving changing the chemical nature of amino acids
citrullination, or deimination, the conversion of arginine to citrulline deamidation, the conversion of glutamine to glutamic acid or asparagine to aspartic acid eliminylation, the conversion to an alkene by beta-elimination of phosphothreonine and phosphoserine, or dehydration of threonine and serine, as well as by decarboxylation of cysteine carbamylation, the conversion of lysine to homocitrulline PTMs involving structural changes
disulfide bridges, the covalent linkage of two cysteine amino acids proteolytic cleavage, cleavage of a protein at a peptide bond racemization of proline by prolyl isomerase [18]
Identification of protein in laboratory
[edit | edit source]The proteins are detected by various method in the laboratory depending upon type of experiment like SDS gel electophoresis, 2D,Western blotting, massspec etc.
SDS-Gel electrophoresis
[edit | edit source]
The sample of proteins are separated by using gel electrophoresis. Separation of proteins may be by isoelectric point (pI), molecular weight, electric charge, or a combination of these factors. The nature of the separation depends on the treatment of the sample and the nature of the gel. This is a very useful way to determine a protein. By far the most common type of gel electrophoresis employs polyacrylamide gels and buffers loaded with sodium dodecyl sulfate (SDS). SDS-PAGE (SDS polyacrylamide gel electrophoresis) maintains polypeptides in a denatured state once they have been treated with strong reducing agents to remove secondary and tertiary structure (e.g. disulfide bonds [S-S] to sulfhydryl groups [SH and SH]) and thus allows separation of proteins by their molecular weight. Sampled proteins become covered in the negatively charged SDS and move to the positively charged electrode through the acrylamide mesh of the gel. Smaller proteins migrate faster through this mesh and the proteins are thus separated according to size (usually measured in kilodaltons, kDa). The concentration of acrylamide determines the resolution of the gel - the greater the acrylamide concentration the better the resolution of lower molecular weight proteins. The lower the acrylamide concentration the better the resolution of higher molecular weight proteins. Proteins travel only in one dimension along the gel for most blots. Samples are loaded into wells in the gel. One lane is usually reserved for a marker or ladder, a commercially available mixture of proteins having defined molecular weights, typically stained so as to form visible, coloured bands. When voltage is applied along the gel, proteins migrate into it at different speeds. These different rates of advancement (different electrophoretic mobilities) separate into bands within each lane.[19]
Setting of transfer in semi dry assembly
[edit | edit source]In order to make the proteins accessible to primary and secondary antibody detection, they are moved from within the gel onto a membrane made of nitrocellulose or polyvinylidene difluoride (PVDF). The membrane is placed on top of the gel, and a stack of filter papers placed on top of that. The entire stack is placed in a buffer solution which moves up the paper by capillary action, bringing the proteins with it. Another method for transferring the proteins is called electroblotting and uses an electric current to pull proteins from the gel into the PVDF or nitrocellulose membrane. The protein move from within the gel onto the membrane while maintaining the organization they had within the gel. As a result of this "blotting" process, the proteins are exposed on a thin surface layer for detection (see below). Both varieties of membrane are chosen for their non-specific protein binding properties (i.e. binds all proteins equally well). Protein binding is based upon hydrophobic interactions, as well as charged interactions between the membrane and protein. Nitrocellulose membranes are cheaper than PVDF, but are far more fragile and do not stand up well to repeated probings.
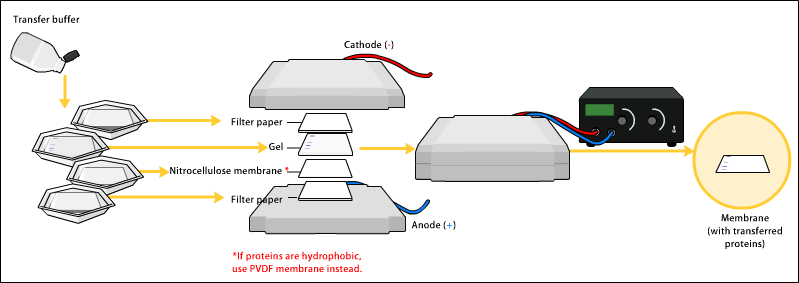
Some laboratories prefer the wet assembly over the semi dry. The uniformity and overall effectiveness of transfer of protein from the gel to the membrane can be checked by staining the membrane with Coomassie Brilliant Blue or Ponceau S dyes. Ponceau S is the more common of the two, due to Ponceau S's higher sensitivity and its water solubility makes it easier to subsequently destain and probe the membrane.
Facts to be remembered
[edit | edit source]The central role of proteins as enzymes in living organisms was however not fully appreciated until 1926, when James B. Sumner showed that the enzyme urease was in fact a protein.
The first protein to be sequenced was insulin, by Frederick Sanger, who won the Nobel Prize for this achievement in 1958.
The first protein structures to be solved were hemoglobin and myoglobin, by Max Perutz and Sir John Cowdery Kendrew, respectively, in 1958.
Till 2009, the Protein Data Bank has over 55,000 atomic-resolution structures of proteins.
The first atomic-resolution structures of proteins were solved by X-ray crystallography in the 1960s and by NMR in the 1980s.
Together with Albert Claude and Christian de Duve, George Emil Palade was awarded the Nobel Prize in Physiology or Medicine, in 1974, for the discovery of the ribosomes.
The Nobel Prize in Chemistry 2009 was awarded to Drs Venkatraman Ramakrishnan, Thomas A. Steitz and Ada E. Yonath "for studies of the structure and function of the ribosome.
Question time
[edit | edit source]1. What do you understand with elongation factors?
2. What are the differences between Eukaryotic and Prokaryotic protein synthesis process?
3. What do you understand with protein synthesis?
4. Define the following terms.
- Kilodalton (kDa)
- Ribosome
- 70S ribosomes vs 80S ribosomes
- Kozak consensus
- Ribosome Recycling Factor
- Peptidyl transfer
5. What is t-RNA and how it is different from m-RNA?
6. Translate the following mRNA codon into one letter and three letter amino acid codes.
AUG ACG CGA GCC UGG CCC GGG GCG CGC AAA ACG GCA GGA ACG ACC AGG UAA
7. Convert the following one letter codes into three letter amino acid codes.
A-P-F-G-C-L-K-Q-M-E-H-I-S-V-X
References
[edit | edit source]- ↑ Protein
- ↑ Protein Biosynthesis
- ↑ Protein
- ↑ Transfer RNA
- ↑ Ribosome
- ↑ http://en.wikipedia.org/wiki/Kozak_consensus_sequence
- ↑ Malys N, McCarthy JEG (2010). "Translation initiation: variations in the mechanism can be anticipated". Cellular and Molecular Life Sciences. doi:10.1007/s00018-010-0588-z.
{{cite journal}}: Unknown parameter|unused_data=ignored (help) - ↑ Structure fo the E. coli protein-coducting channel bound to at translating ribosome, K. Mitra, et al. Nature (2005), vol 438, p 318
- ↑ http://en.wikipedia.org/wiki/Prokaryotic_translation
- ↑ Hirokawa et al. (2006) "The Ribosome Recycling Step: Consensus or Controversy?". Trends in Biochemical Sciences Vol. 31(3), 143-149.
- ↑ Eukaryotic initiation factor
- ↑ http://en.wikipedia.org/wiki/Eukaryotic_initiation_factor
- ↑ http://en.wikipedia.org/wiki/Eukaryotic_initiation_factor
- ↑ Eukaryotic translation
- ↑ http://en.wikipedia.org/w/index.php?title=Eukaryotic_translation&oldid=429826292
- ↑ Protein structure
- ↑ http://en.wikipedia.org/w/index.php?title=Protein_secondary_structure&oldid=425646887
- ↑ Posttranslational modification
- ↑ http://en.wikipedia.org/w/index.php?title=Western_blot&oldid=429925576
