Data Mining Algorithms In R/Dimensionality Reduction/Principal Component Analysis
Introduction
[edit | edit source]This chapter presents the Principal Component Analysis (PCA) technique as well as its use in R project for statistical computing. First we will introduce the technique and its algorithm, second we will show how PCA was implemented in the R language and how to use it. Finally, we will present an example of an application of the technique in a data mining scenario. In the end of the chapter you will find references for further information.
Principal Component Analysis
[edit | edit source]PCA is a dimensionality reduction method in which a covariance analysis between factors takes place. The original data is remapped into a new coordinate system based on the variance within the data. PCA applies a mathematical procedure for transforming a number of (possibly) correlated variables into a (smaller) number of uncorrelated variables called principal components. The first principal component accounts for as much of the variability in the data as possible, and each succeeding component accounts for as much of the remaining variability as possible.
PCA is useful when there is data on a large number of variables, and (possibly) there is some redundancy in those variables. In this case, redundancy means that some of the variables are correlated with one another. And because of this redundancy, PCA can be used to reduce the observed variables into a smaller number of principal components that will account for most of the variance in the observed variables.
PCA is recommended as an exploratory tool to uncover unknown trends in the data. The technique has found application in fields such as face recognition and image compression, and is a common technique for finding patterns in data of high dimension.
Algorithm
[edit | edit source]The PCA algorithm consists of 5 main steps:
- Subtract the mean: subtract the mean from each of the data dimensions. The mean subtracted is the average across each dimension. This produces a data set whose mean is zero.
- Calculate the covariance matrix:
- where is a matrix which each entry is the result of calculating the covariance between two separate dimensions.
- Calculate the eigenvectors and eigenvalues of the covariance matrix.
- Choose components and form a feature vector: once eigenvectors are found from the covariance matrix, the next step is to order them by eigenvalue, highest to lowest. So that the components are sorted in order of significance. The number of eigenvectors that you choose will be the number of dimensions of the new data set. The objective of this step is construct a feature vector (matrix of vectors). From the list of eigenvectors take the eigenvectors selected and form a matrix with them in the columns:
- FeatureVector = (eig_1, eig_2, ..., eig_n)
- Derive the new data set. Take the transpose of the FeatureVector and multiply it on the left of the original data set, transposed:
- FinalData = RowFeatureVector x RowDataAdjusted
- where RowFeatureVector is the matrix with the eigenvectors in the columns transposed (the eigenvectors are now in the rows and the most significant are in the top) and RowDataAdjusted is the mean-adjusted data transposed (the data items are in each column, with each row holding a separate dimension).
Implementation
[edit | edit source]We choose princomp method from stats package for this tutorial.
- R package: stats
- Method: princomp
- Documentation: princomp
princomp is a generic method with "formula" and "default" methods from stats package which performs a principal components analysis on the given numeric data matrix and returns the results as an object of class princomp.
Usage
princomp(x, ...)
- S3 method for class 'formula:
princomp(formula, data = NULL, subset, na.action, ...)
- Default S3 method:
princomp(x, cor = FALSE, scores = TRUE, covmat = NULL, subset = rep(TRUE, nrow(as.matrix(x))), ...)
- S3 method for class 'princomp':
predict(object, newdata, ...)
Arguments
- formula
- a formula with no response variable, referring only to numeric variables.
- data
- an optional data frame containing the variables in the formula formula. By default the variables are taken from environment(formula)
- subset
- an optional vector used to select rows (observations) of the data matrix x.
- na.action
- a function which indicates what should happen when the data contain NAs. The default is set by the na.action setting of options, and is na.fail if that is unset. The ‘factory-fresh’ default is na.omit.
- x
- a numeric matrix or data frame which provides the data for the principal components analysis.
- cor
- a logical value indicating whether the calculation should use the correlation matrix or the covariance matrix. (The correlation matrix can only be used if there are no constant variables.)
- scores
- a logical value indicating whether the score on each principal component should be calculated.
- covmat
- a covariance matrix, or a covariance list as returned by cov.wt (and cov.mve or cov.mcd from package MASS). If supplied, this is used rather than the covariance matrix of x.
- ...
- arguments passed to or from other methods. If x is a formula one might specify cor or scores.
- object
- Object of class inheriting from "princomp"
- newdata
- An optional data frame or matrix in which to look for variables with which to predict. If omitted, the scores are used. If the original fit used a formula or a data frame or a matrix with column names, newdata must contain columns with the same names. Otherwise it must contain the same number of columns, to be used in the same order.
Value
The princomp method returns a list with class "princomp" containing the following components:
- sdev
- a formula with no response variable, referring only to numeric variables.
- loadings
- the matrix of variable loadings (i.e., a matrix whose columns contain the eigenvectors).
- center
- the means that were subtracted.
- scale
- the scalings applied to each variable.
- n.obs
- the number of observations.
- scores
- if scores = TRUE, the scores of the supplied data on the principal components. These are non-null only if x was supplied, and if covmat was also supplied if it was a covariance list. For the formula method, napredict() is applied to handle the treatment of values omitted by the na.action.
- call
- the matched call.
- na.action
- if relevant.
Visualization
[edit | edit source]The print method for these objects prints the results in a nice format and the plot method produces a screen plot. There is also a biplot method.
Examples
require(graphics)
summary(pc.cr <- princomp(USArrests, cor = TRUE))
loadings(pc.cr)
plot(pc.cr)
biplot(pc.cr)
Case Study
[edit | edit source]To illustrate the PCA technique for dimensionality reduction, a simple case study will be shown.
Scenario
[edit | edit source]In the field of information retrieval (IR), queries and documents can be represented in a vector space.
Generally, several features are used to describe a document retrieved by a query such as TF-IDF and PageRank measures.
Occasionally, it can be necessary to visualize documents in a 2-dimensional space. To do this, PCA could be used.
Dataset
[edit | edit source]More recently, Microsoft released the LETOR benchmark data sets for research on LEarning TO Rank, which contains standard features, relevance judgments, data partitioning, evaluation tools, and several baselines. LETOR contains several datasets for ranking settings derived from the two query sets and the Gov2 web page collection. The 5-fold cross validation strategy is adopted and the 5-fold partitions are included in the package. In each fold, there are three subsets for learning: training set, validation set and testing set. The datasets can be downloaded from LETOR site.
A typical document in a LETOR dataset is described as follows:
0 qid:1 1:1.000000 2:1.000000 3:0.833333 4:0.871264 5:0 6:0 7:0 8:0.941842 9:1.000000 10:1.000000 11:1.000000 12:1.000000 13:1.000000 14:1.000000 15:1.000000 16:1.000000 17:1.000000 18:0.719697 19:0.729351 20:0 21:0 22:0 23:0.811565 24:1.000000 25:0.972730 26:1.000000 27:1.000000 28:0.922374 29:0.946654 30:0.938888 31:1.000000 32:1.000000 33:0.711276 34:0.722202 35:0 36:0 37:0 38:0.798002 39:1.000000 40:1.000000 41:1.000000 42:1.000000 43:0.959134 44:0.963919 45:0.971425 #docid = 244338
Execution
[edit | edit source]LETOR files can not be imported 'as is', but using the following script, it is possible to convert LETOR format to other accepted by R, as can be seen below:
cat train.txt | grep "qid:1 " | gawk '{ printf("doc_"$50); for (i=3;i<=47;i++) { split($i,a,":"); printf(", "a[2]); } printf("\n"); }' > letor.data
After that, 'letor.data' will be seen as follows:
doc_244338, 1.000000, 1.000000, 0.833333, 0.871264, 0, 0, 0, 0.941842, 1.000000, 1.000000, 1.000000, 1.000000, 1.000000, 1.000000, 1.000000, 1.000000, 1.000000, 0.719697, 0.729351, 0, 0, 0, 0.811565, 1.000000, 0.972730, 1.000000, 1.000000, 0.922374, 0.946654, 0.938888, 1.000000, 1.000000, 0.711276, 0.722202, 0, 0, 0, 0.798002, 1.000000, 1.000000, 1.000000, 1.000000, 0.959134, 0.963919, 0.971425
Now, it is possible to load 'letor.data' file to R and run PCA to plot documents in a 2-dimensional space, as follows:
data=read.table("letor.data",sep=",")
summary(pc.cr <- princomp(data[,2:46]))
loadings(pc.cr)
library(lattice)
pc.cr$scores
pca.plot <- xyplot(pc.cr$scores[,2] ~ pc.cr$scores[,1])
pca.plot$xlab <- "First Component"
pca.plot$ylab <- "Second Component"
pca.plot
Results
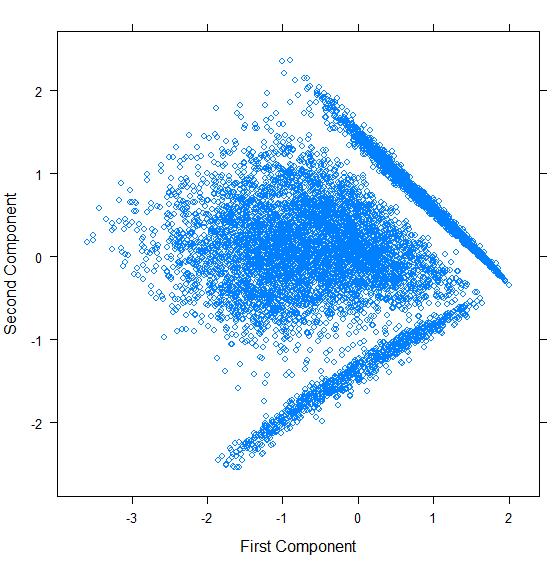
[edit | edit source]The script above generates the following chart:
Analysis
[edit | edit source]As we can see, despite documents being represented by multiple features (>45), PCA was able to find 2 principal components that were used to plot all documents in a 2-dimensional chart.
References
[edit | edit source]- ^ Mardia, K. V., J. T. Kent and J. M. Bibby (1979). "Multivariate Analysis", London: Academic Press.
- ^ Venables, W. N. and B. D. Ripley (2002). "Modern Applied Statistics with S", Springer-Verlag.
- Principal Components Analysis and Redundancy Analysis Lab, Montana State University. Link.
- Visualising and exploring multivariate datasets using singular value decomposition and self organising maps from Bioinformatics Zen. Link.