Methods and Concepts in the Life Sciences/DNA Sequencing
DNA sequencing
[edit | edit source]Classical methods
[edit | edit source]Maxam-Gilbert sequencing
[edit | edit source]Allan Maxam and Walter Gilbert published a DNA sequencing method in 1977 based on chemical modification of DNA and subsequent cleavage at specific bases. Also known as chemical sequencing, this method allowed purified samples of double-stranded DNA to be used without further cloning. This method's use of radioactive labeling and its technical complexity discouraged extensive use after refinements in the Sanger methods had been made.
Maxam-Gilbert sequencing requires radioactive labeling at one 5' end of the DNA and purification of the DNA fragment to be sequenced. Chemical treatment then generates breaks at a small proportion of one or two of the four nucleotide bases in each of four reactions (G, A+G, C, C+T). The concentration of the modifying chemicals is controlled to introduce on average one modification per DNA molecule. Thus a series of labeled fragments is generated, from the radiolabeled end to the first "cut" site in each molecule. The fragments in the four reactions are electrophoresed side by side in denaturing acrylamide gels for size separation. To visualize the fragments, the gel is exposed to X-ray film for autoradiography, yielding a series of dark bands each corresponding to a radiolabeled DNA fragment, from which the sequence may be inferred.

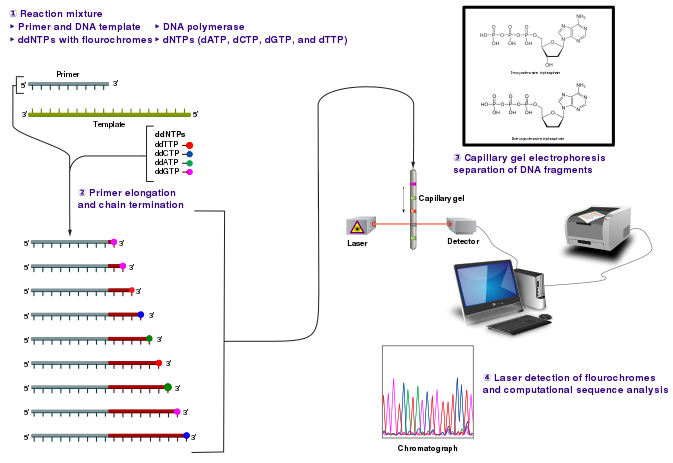
Sanger sequencing
[edit | edit source]
The chain-termination method developed by Frederick Sanger and coworkers in 1977 soon became the method of choice, owing to its relative ease and reliability. When invented, the chain-terminator method used fewer toxic chemicals and lower amounts of radioactivity than the Maxam and Gilbert method. Because of its comparative ease, the Sanger method was soon automated and was the method used in the first generation of DNA sequencers.
The classical chain-termination method requires a single-stranded DNA template, a DNA primer, a DNA polymerase, normal deoxynucleosidetriphosphates (dNTPs), and modified di-deoxynucleotidetriphosphates (ddNTPs), the latter of which terminate DNA strand elongation. These chain-terminating nucleotides lack the 3'-OH group required for the formation of a phosphodiester bond between two nucleotides, causing DNA polymerase to cease extension of DNA when a ddNTP is incorporated.
The DNA sample is divided into four separate sequencing reactions, containing all four of the standard dNTPs and the DNA polymerase. To each reaction is added only one of the four dideoxynucleotides (ddATP, ddGTP, ddCTP, or ddTTP). Following rounds of template DNA extension from the bound primer, the resulting DNA fragments are heat denatured and separated by size using gel electrophoresis. In the original publication of 1977, Sanger used radioactively labeled dATP for detection of the bands via autoradiography.
The image to the right shows an X-ray film which was exposed a sequencing gel, and the dark bands correspond to DNA fragments of different lengths. A dark band in a lane indicates a DNA fragment that is the result of chain termination after incorporation of a certain ddNTP. The relative positions of the different bands among the four lanes, from bottom to top, are then used to read the DNA sequence.
Technical variations of chain-termination sequencing include using a labelled primer, which can be either radioactive or contain a fluorescent dye. Dye-primer sequencing facilitates reading in an optical system for faster and more economical analysis and automation.
Cycle sequencing
[edit | edit source]Cycle sequencing combines Sanger sequencing and PCR to sequence very small amounts of DNA. The sequencing reaction takes place in a thermocycler, where the repeated denaturation, annealing and DNA synthesis by a thermostable polymerase leads to an amplification of the chain-termination products. In contrast to normal PCR, this amplification is not exponential but linear, because only one primer is used.
Dye-terminator sequencing
[edit | edit source]
Dye-terminator sequencing utilizes labelling of the chain terminator ddNTPs, which permits sequencing in a single reaction, rather than four reactions as in the labelled-primer method. In dye-terminator sequencing, each of the four ddNTP chain terminators is labelled with fluorescent dyes, each of which emit light at different wavelengths.
Owing to its greater expediency and speed, dye-terminator sequencing is now the mainstay in automated sequencing. Its limitations include dye effects due to differences in the incorporation of the dye-labelled chain terminators into the DNA fragment, resulting in unequal peak heights and shapes in the electronic DNA sequence trace chromatogram after capillary electrophoresis.
This problem has been addressed with the use of modified DNA polymerase enzyme systems and dyes that minimize incorporation variability, as well as methods for eliminating "dye blobs". The dye-terminator sequencing method, along with automated high-throughput DNA sequence analyzers, is now being used for the vast majority of sequencing projects.
Limitations
[edit | edit source]Common challenges of DNA sequencing with the Sanger method include poor quality in the first 15-40 bases of the sequence due to primer binding and deteriorating quality of sequencing traces after 700-900 bases. Current methods can directly sequence only relatively short (300-1000 nucleotides long) DNA fragments in a single reaction. The main obstacle to sequencing DNA fragments above this size limit is insufficient power of separation for resolving large DNA fragments that differ in length by only one nucleotide.

Advanced methods
[edit | edit source]Pyrosequencing
[edit | edit source]The pyrosequencing method was developed in 1996 and is based on detecting the activity of DNA polymerase with a chemiluminescent enzyme. Essentially, the method allows sequencing of a single strand of DNA by synthesizing the complementary strand along it, one base pair at a time, and detecting which base was actually added at each step. The template DNA is immobile, and solutions of A, C, G, and T nucleotides are sequentially added and removed from the reaction. Light is produced only when the nucleotide solution complements the first unpaired base of the template. The sequence of solutions which produce chemiluminescent signals allows the determination of the sequence of the template.
An ssDNA template is hybridized to a sequencing primer and incubated with the enzymes DNA polymerase, ATP sulfurylase, luciferase and apyrase, and with the substrates adenosine 5´ phosphosulfate (APS) and luciferin.
- The addition of one of the four deoxynucleoside triphosphates (dNTPs) initiates the second step. DNA polymerase incorporates the correct, complementary dNTPs onto the template. This incorporation releases pyrophosphate (PPi) stoichiometrically.
- ATP sulfurylase quantitatively converts PPi to ATP in the presence of adenosine 5´ phosphosulfate. This ATP acts as a substrate for the luciferase-mediated conversion of luciferin to oxyluciferin that generates visible light in amounts that are proportional to the amount of ATP. The light produced in the luciferase-catalyzed reaction is detected by a camera and analyzed in a pyrogram.
- Unincorporated nucleotides and ATP are degraded by the apyrase, and the reaction can restart with another nucleotide.
Currently, a limitation of the method is that the lengths of individual reads of DNA sequence are in the neighborhood of 300-500 nucleotides, shorter than the 800-1000 obtainable with chain termination methods (e.g. Sanger sequencing). This can make the process of genome assembly more difficult, particularly for sequences containing a large amount of repetitive DNA. As of 2007, pyrosequencing is most commonly used for resequencing or sequencing of genomes for which the sequence of a close relative is already available.
The templates for pyrosequencing can be made both by solid phase template preparation (streptavidin-coated magnetic beads) and enzymatic template preparation (apyrase+exonuclease). Thus, pyrosequencing is differentiated into two types; namely Solid Phase Pyrosequencing and Liquid Phase Pyrosequencing.
Next-generation sequencing
[edit | edit source]The high demand for low-cost sequencing has driven the development of high-throughput sequencing (or next-generation sequencing) technologies that parallelize the sequencing process, producing thousands or millions of sequences concurrently. High-throughput sequencing technologies are intended to lower the cost of DNA sequencing beyond what is possible with standard dye-terminator methods. In ultra-high-throughput sequencing as many as 500,000 sequencing-by-synthesis operations may be run in parallel.
454 sequencing
[edit | edit source]454 Sequencing uses a large-scale parallel pyrosequencing system capable of sequencing roughly 400-600 megabases of DNA per 10-hour run.
Genomic DNA is fractionated into smaller fragments (300-800 base pairs) and polished (made blunt at each end). Short adaptors are then ligated onto the ends of the fragments. These adaptors provide priming sequences for both amplification and sequencing of the sample-library fragments. One adaptor (Adaptor B) contains a 5'-biotin tag for immobilization of the DNA library onto streptavidin-coated beads. After nick repair, the non-biotinylated strand is released and used as a single-stranded template DNA (sstDNA) library. The sstDNA library is assessed for its quality and the optimal amount (DNA copies per bead) needed for PCR is determined by titration.
The sstDNA library is immobilized onto beads. The beads containing a library fragment carry a single sstDNA molecule. The bead-bound library is emulsified with the amplification reagents in a water-in-oil mixture. Each bead is captured within its own microreactor where PCR amplification occurs. This results in bead-immobilized, clonally amplified DNA fragments.
Single-stranded template DNA library beads are added to the DNA Bead Incubation Mix (containing DNA polymerase) and are layered with Enzyme Beads (containing sulfurylase and luciferase) onto a PicoTiterPlate device. The device is centrifuged to deposit the beads into the wells. The layer of Enzyme Beads ensures that the DNA beads remain positioned in the wells during the sequencing reaction. The bead-deposition process is designed to maximize the number of wells that contain a single amplified library bead.
The loaded PicoTiterPlate is placed into the Genome Sequencer FLX Instrument. The fluidics sub-system delivers sequencing reagents (containing buffers and nucleotides) across the wells of the plate. The four DNA nucleotides are added sequentially in a fixed order across the PicoTiterPlate during a sequencing run. During the nucleotide flow, millions of copies of DNA bound to each of the beads are sequenced in parallel. When a nucleotide complementary to the template strand is added into a well, the polymerase extends the existing DNA strand by adding a nucleotide. Addition of one (or more) nucleotide(s) generates a light signal that is recorded by the CCD camera in the instrument. The signal strength is proportional to the number of nucleotides; for example, homopolymer stretches, incorporated in a single nucleotide flow generate a greater signal than single nucleotides. However, the signal strength for homopolymer stretches is linear only up to eight consecutive nucleotides after which the signal falls-off rapidly. Data are stored in standard flowgram format (SFF) files for downstream analysis.
In late March 2007, Roche Diagnostics announced an agreement to purchase 454 Life Sciences for US$154.9 million. In October 2013, Roche announced that it will shut down 454, and stop supporting the platform by mid-2016.
Illumina sequencing
[edit | edit source]
Solexa, now part of Illumina, was founded in 1998 and developed a sequencing-by-synthesis technology based on reversible terminators. In this method, the DNA is first sheared and two adapters are ligated to the ends of the fragments. Single-stranded, adapter-ligated fragments are then bound randomly to the surface of a flow cell and amplified by bridge amplification: The flow cell is coated with oligonucleotides that correspond to the adapters. Therefore, the free end of the fragments can “bridge” to a complementary oligo on the surface, which then serves as a primer for a DNA polymerase. Denaturation of the newly synthesized double-stranded bridge leaves two single-stranded fragments which are attached to the surface. Repeated extension and denaturation generates millions of unique clusters (or polonies, for polymerase generated colonies) across the flow cell. In the next step, these colonies can be sequenced using reversible terminators. These are nucleotides which are chemically blocked at the 3'‑OH group. Furthermore, the four types of nucleotides carry different fluorescent labels. For the first cycle of sequencing, all four nucleotides, a primer and a DNA polymerase are added to the flow cell. At each template, a single nucleotide can be incorporated, the remaining are washed away. Now the fluorescent labels are excited with a laser and a high resolution image of the whole flow cell is taken. Any signal above background identifies the physical location of a cluster and the fluorescent emission identifies which of the four bases was incorporated. Next, the dye and the terminal 3' blocker are removed and the four reversible terminators and the polymerase are added to begin the next cycle. In this way, the sequence of the fragments can be determined one base at a time. In the end, millions of reads can be aligned to yield the sequence of the original DNA.
Illumina is currently the market leader in the field of next-generation sequencing machines. It offers several versions of its sequencers, of which the HiSeq is the most powerful and commonly used to sequence large genomes. According to Illumina, the HiSeq X Ten is the first sequencing platform to break the $1000 barrier for a human genome. It can produce up to 3 billion reads per flow cell, with a maximum read length of 150 bp from both ends of each template. The run time is up to three days. The MiSeq is a faster and less expensive alternative, with 25 million reads per flow cell and a maximum read length of 300 bp from each end.
Ion Torrent semiconductor sequencing
[edit | edit source]Ion Torrent Systems Inc. (now owned by Life Technologies) developed a system based on using standard sequencing chemistry, but with a novel, semiconductor based detection system. This method of sequencing is based on the detection of hydrogen ions that are released during the polymerisation of DNA, as opposed to the optical methods used in other sequencing systems. A microwell containing a template DNA strand to be sequenced is flooded with a single type of nucleotide. If the introduced nucleotide is complementary to the leading template nucleotide it is incorporated into the growing complementary strand. This causes the release of a hydrogen ion that triggers a hypersensitive ion sensor, which indicates that a reaction has occurred. If homopolymer repeats are present in the template sequence multiple nucleotides will be incorporated in a single cycle. This leads to a corresponding number of released hydrogens and a proportionally higher electronic signal.

Single molecule real time (SMRT) sequencing
[edit | edit source]SMRT sequencing is based on the sequencing-by-synthesis approach. The DNA is synthesized in zero-mode wave-guides (ZMWs) – small well-like containers with the capturing tools located at the bottom of the well. The sequencing is performed with an unmodified polymerase (attached to the ZMW bottom) and fluorescently labeled nucleotides flowing freely in the solution. The wells are constructed in such a way that only the fluorescence occurring at the bottom of the well is detected. The fluorescent label is detached from the nucleotide upon its incorporation into the DNA strand, leaving an unmodified DNA strand. According to Pacific Biosciences, the SMRT technology developer, this methodology allows detection of nucleotide modifications (such as cytosine methylation). This happens through the observation of polymerase kinetics. This approach allows reads of 20,000 nucleotides or more, with average read lengths of 5 kilobases.
Nanopore sequencing
[edit | edit source]This method is based on the readout of electrical signals occurring at nucleotides passing by alpha-hemolysin pores covalently bound with cyclodextrin. The DNA passing through the nanopore changes its ion current. This change is dependent on the shape, size and length of the DNA sequence. Each type of the nucleotide blocks the ion flow through the pore for a different period of time. The method has a potential of development as it does not require modified nucleotides, however single nucleotide resolution is not yet available.
Two main areas of nanopore sequencing in development are solid state nanopore sequencing, and protein based nanopore sequencing. Protein nanopore sequencing utilizes membrane protein complexes such as ∝‑Hemolysin and MspA (Mycobacterium Smegmatis Porin A), which show great promise given their ability to distinguish between individual and groups of nucleotides. Solid-state nanopore sequencing utilizes synthetic materials such as silicon nitride and aluminum oxide and it is preferred for its superior mechanical ability and thermal and chemical stability. The fabrication method is essential for this type of sequencing given that the nanopore array can contain hundreds of pores with diameters smaller than eight nanometers.
The concept originated from the idea that single stranded DNA or RNA molecules can be electrophoretically driven in a strict linear sequence through a biological pore that can be less than eight nanometers, and can be detected given that the molecules release an ionic current while moving through the pore. The pore contains a detection region capable of recognizing different bases, with each base generating various time specific signals corresponding to the sequence of bases as they cross the pore which are then evaluated. When implementing this process it is important to note that precise control over the DNA transport through the pore is crucial for success. Various enzymes such as exonucleases and polymerases have been used to moderate this process by positioning them near the pore’s entrance.
Sequencing strategies
[edit | edit source]Primer walking
[edit | edit source]Primer walking is a method for sequencing DNA fragments between 1.3 and 7 kilobases. Such fragments are too long to be sequenced in a single sequence read using the chain termination method. This method works by dividing the long sequence into several consecutive short ones. The DNA of interest may be a plasmid insert, a PCR product or a fragment representing a gap when sequencing a genome.
The fragment is first sequenced as if it were a shorter fragment — sequencing will be performed from each end using either universal primers or primers designated by the customer. This should identify the first 1000 (approx.) bases. In order to completely sequence the region of interest, design and synthesis of new primers — complementary to the final 20 bases of the known sequence — is necessary to obtain contiguous sequence information.
That way, the short part of the long DNA that is sequenced keeps "walking" along the sequence. The method can be used to sequence entire chromosomes (thus, chromosome walking).
Shotgun sequencing
[edit | edit source]In shotgun sequencing, DNA is broken up randomly into numerous small segments, which are sequenced to obtain reads. Multiple overlapping reads for the target DNA are obtained by performing several rounds of this fragmentation and sequencing. Computer programs then use the overlapping ends of different reads to assemble them into a continuous sequence.
The classical shotgun sequencing was based on the Sanger sequencing method: this was the most advanced technique for sequencing genomes from about 1995–2005. The shotgun strategy is still applied today, however using next-generation sequencing. These technologies produce shorter reads (anywhere from 25–500bp) but many hundreds of thousands or millions of reads in a relatively short time (on the order of a day). This results in high coverage, but the assembly process is much more computationally expensive. These technologies are vastly superior to Sanger sequencing due to the high volume of data and the relatively short time it takes to sequence a whole genome.
References
[edit | edit source]- Mardis, E.R., 2008. Next-generation DNA sequencing methods. Annu. Rev. Genomics Hum. Genet. 9, 387–402.
- Metzker, M.L., 2010. Sequencing technologies - the next generation. Nat. Rev. Genet. 11, 31–46.
- Sanger, F., Coulson, A.R., 1975. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J. Mol. Biol. 94, 441–8.
- Sanger, F., Nicklen, S., Coulson, A.R., 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U. S. A. 74, 5463‑7.
