Methods and Concepts in the Life Sciences/PCR
PCR
[edit | edit source]
The polymerase chain reaction (PCR) is used to amplify a piece of DNA across several orders of magnitude, generating thousands to millions of copies of a particular DNA sequence.
Developed in 1983 by Kary Mullis, PCR is now a common and often indispensable technique used in medical and biological research labs for a variety of applications. In 1993, Mullis was awarded the Nobel Prize in Chemistry along with Michael Smith for his work on PCR.
The method relies on thermal cycling, consisting of cycles of repeated heating and cooling of the reaction for DNA melting and enzymatic replication of the DNA. Primers containing sequences complementary to the target region along with a heat-stable DNA polymerase are key components to enable selective and repeated amplification. As PCR progresses, the DNA generated is itself used as a template for replication, setting in motion a chain reaction in which the DNA template is exponentially amplified. PCR can be extensively modified to perform a wide array of genetic manipulations.
Materials
[edit | edit source]

PCR is used to amplify a specific region of a DNA strand (the DNA target). Most PCR methods typically amplify DNA fragments of between 0.1 and 10 kilo base pairs (kbp), although some techniques allow for amplification of fragments up to 40 kbp in size. The amount of amplified product is determined by the available substrates in the reaction, which become limiting as the reaction progresses.
A basic PCR set up requires several components and reagents. These components include:
- DNA template that contains the DNA region to be amplified.
- Two primers that are complementary to the 3' ends of each of the sense and anti-sense strand of the DNA target.
- A thermostable DNA polymerase
- Deoxynucleoside triphosphates (dNTPs)
- Buffer solution, providing a suitable chemical environment for optimum activity and stability of the DNA polymerase.
The PCR is commonly carried out in a reaction volume of 10–200 μl in small reaction tubes (0.2–0.5 ml volumes) in a thermal cycler. The thermal cycler heats and cools the reaction tubes to achieve the temperatures required at each step of the reaction (see below). Many modern thermal cyclers make use of the Peltier effect, which permits both heating and cooling of the block holding the PCR tubes simply by reversing the electric current. Thin-walled reaction tubes permit favorable thermal conductivity to allow for rapid thermal equilibration. Most thermal cyclers have heated lids to prevent condensation at the top of the reaction tube. Older thermocyclers lacking a heated lid require a layer of oil on top of the reaction mixture or a ball of wax inside the tube.
Procedure
[edit | edit source]
Typically, PCR consists of a series of 20-40 repeated temperature changes, with each cycle commonly consisting of three discrete temperature steps. The cycling is often preceded by a single temperature step at a high temperature (>90 °C), and followed by one hold at the end for final product extension or brief storage. The temperatures used and the length of time they are applied in each cycle depend on a variety of parameters. These include the polymerase used for DNA synthesis, the concentration of divalent ions and dNTPs in the reaction, and the melting temperature (Tm) of the primers.
- Initialization (Only required for DNA polymerases that require heat activation by hot-start PCR.): This step consists of heating the reaction to a temperature of 94–96 °C (or 98 °C if extremely thermostable polymerases are used), which is held for 1–9 minutes.
- Denaturation: This step is the first regular cycling event and consists of heating the reaction to 94–98 °C for 20–30 seconds. It causes the strands of the DNA template to separate by disrupting the hydrogen bonds between complementary bases, yielding single-stranded DNA molecules.
- Annealing: The reaction temperature is lowered to 50–65 °C for 20–40 seconds, allowing annealing of the primers to the single-stranded DNA template. This temperature needs to be low enough to allow for hybridization of the primer to the strand, but high enough in order for the hybridization to be specific, i.e. the primer should only bind to a perfectly complementary part of the template. If the temperature is too low, the primer could bind imperfectly. If it is too high, the primer might not bind. Typically, the annealing temperature is about 3–5 °C below the Tm of the primers used. Stable DNA–DNA hydrogen bonds are only formed when the primer sequence very closely matches the template sequence. The polymerase binds to the primer-template hybrid and begins DNA formation.
- Elongation: The temperature at this step depends on the DNA polymerase used; Taq polymerase has its optimum activity temperature at 75–80 °C, and commonly a temperature of 72 °C is used with this enzyme. At this step the DNA polymerase synthesizes a new DNA strand complementary to the DNA template strand by adding dNTPs that are complementary to the template in 5' to 3' direction, condensing the 5'-phosphate group of the dNTPs with the 3'-hydroxyl group at the end of the nascent DNA strand. The extension time depends both on the DNA polymerase used and on the length of the DNA fragment to be amplified. As a rule-of-thumb, at its optimum temperature, the DNA polymerase will polymerize a thousand bases per minute. Under optimum conditions, i.e., if there are no limitations due to limiting substrates or reagents, at each extension step, the amount of DNA target is doubled. After elongation, the DNA is denatured again and the newly synthesized fragments can serve as templates in the next cycle, leading to exponential amplification of the specific DNA fragment.
- Final elongation: This single step is occasionally performed at a temperature of 70–74 °C (this is the temperature needed for optimal activity for most polymerases used in PCR) for 5–15 minutes after the last PCR cycle to ensure that any remaining single-stranded DNA is fully extended.
- Final hold: This step at 4–15 °C for an indefinite time may be employed for short-term storage of the reaction.
Primer design
[edit | edit source]Good primers are essential for a successful PCR reaction. Several factors should be taken into account when designing them:
- In general, primers should have a length of 18–30 nucleotides.
- Try to make the melting temperature (Tm) of the primers between 65°C and 75°C, and within 5°C of each other.
- Aim for a GC content between 40 and 60%.
- Use a G or C at the 3’-end of each primer (“GC clamp”) to promote binding. However, 3 of more G or C bases at this end may stabilize nonspecific annealing of the primer.
- Primers should not be self-complementary or complementary to the other primer in the reaction, otherwise they can form dimers. Especially the 3'-end of the primer is critical.
- Primers should not form stable secondary structures.
- Try to avoid runs of 4 or more of one base, or dinucleotide repeats.
- Primers should be compared to the template sequence to detect secondary binding sites.
- When adding a restriction site to the 5’ end of a primer, add 3 to 4 additional nucleotides to allow for efficient cutting.
The following formula can be used as a rule of thumb to estimate the melting temperature of a primer:
Since GC base pairs are more stable than AT pairs, it is assumed that each G or C increases the melting temperature by 4 °C, whereas an A or T only contributes 2 °C. There are several tools which use more sophisticated algorithms to calculate the Tm:
- OligoAnalyzer calculates the Tm and can also check for dimers and hairpins.
- Oligonucleotide Properties Calculator calculates the Tm using different algorithms.
- NEB Tm Calculator takes the polymerase into account.
- AmplifX evaluates several properties of primers.
Primers can be designed manually or using software which suggests primer pairs for a given template sequence based on several predetermined parameters.
- Primer3 designs primers and gives the user significant control over the nature of the primers.
- Primer-BLAST uses Primer3 to design primers and then automatically analyzes their specificity using BLAST.
These are only some commonly used examples out of the huge variety of tools which can be used to design and analyze primers. Some tools are optimized for special applications, such as Real-time PCR. The following websites provide an overview of primer designing tools:
Real-time PCR
[edit | edit source]Real-time or quantitative PCR is variant of PCR which is used to amplify and simultaneously detect or quantify a DNA molecule. The procedure follows the general principle of polymerase chain reaction; its key feature is that the amplified DNA is detected as the reaction progresses in "real time". This is a new approach compared to standard PCR, where the product of the reaction is detected at its end. Two common methods for the detection of products in quantitative PCR are:
- Non-specific fluorescent dyes that intercalate with any double-stranded DNA.
- Sequence-specific DNA probes consisting of oligonucleotides that are labelled with a fluorescent reporter.
The MIQE guidelines (minimum information for publication of quantitative real-time PCR experiments) propose that the abbreviation qPCR be used for quantitative real-time PCR and that RT-qPCR be used for reverse transcription–qPCR. The acronym "RT-PCR" commonly denotes reverse transcription polymerase chain reaction and not real-time PCR, but not all authors adhere to this convention.
Detection methods
[edit | edit source]Fluorescent dyes
[edit | edit source]The easiest way of detecting and quantifying PCR products is the use of DNA-binding dyes such as ethidium bromide or SYBR Green. The fluorescence intensity of these fluorophores increases when they are bound to DNA, which means that the accumulation of DNA in the course of a PCR correlates with an increasing fluorescence. The measurement takes place at the end of elongation in each cycle.
The disadvantage of this method is the low specificity. It is not possible to distinguish between different PCR products and unspecific products such as primer dimers can interfere with the accurate quantification of the target sequence.
Fluorescent probes
[edit | edit source]Fluorescent reporter probes detect only the DNA containing the probe sequence; therefore, use of the reporter probe significantly increases specificity, and enables quantification even in the presence of non-specific DNA amplification. Fluorescent probes can be used in multiplex assays—for detection of several genes in the same reaction—based on specific probes with different-coloured labels, provided that all targeted genes are amplified with similar efficiency.
These methods rely on Förster resonance energy transfer (FRET) to detect whether two fluorochromes are in spatial proximity or not. FRET is a mechanism of energy transfer between two light-sensitive molecules. A donor chromophore, initially in its electronic excited state, may transfer energy to an acceptor chromophore through nonradiative dipole–dipole coupling. The efficiency of this energy transfer is inversely proportional to the sixth power of the distance between donor and acceptor, making FRET extremely sensitive to small changes in distance. Measurements of FRET efficiency can therefore be used to determine if two fluorophores are within a certain distance of each other.
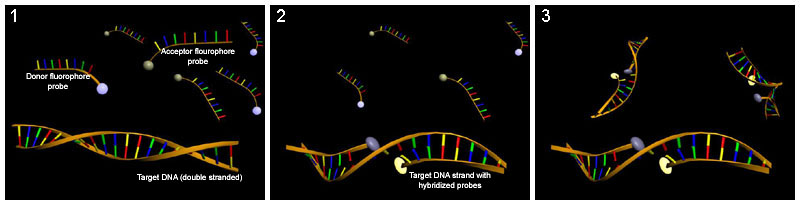
LightCycler probes
[edit | edit source]LightCycler probes consist of two oligonucleotides which are complementary to adjacent regions of the target sequence. One probe carries a FRET donor and the other a FRET acceptor. When both oligonucleotides anneal to the target, FRET can occur and the fluorescence intensity can be used for quantification.
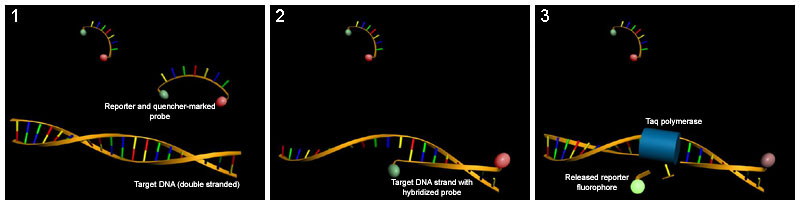
TaqMan probes
[edit | edit source]This method relies on an oligonucleotide with a fluorescent reporter at one end and a quencher of fluorescence at the opposite end. As long as the probe is intact, the close proximity of the reporter to the quencher prevents detection of its fluorescence. The probe anneals between the two primers and is degraded by the 5'-3' exonuclease activity of Taq polymerase during elongation. This breaks the reporter-quencher proximity and thus allows unquenched emission of fluorescence, which can be detected after excitation with a laser. An increase in the product targeted by the reporter probe at each PCR cycle therefore causes a proportional increase in fluorescence due to the breakdown of the probe and release of the reporter.
Quantification
[edit | edit source]
Unlike end point PCR (conventional PCR) real time PCR allows quantification of the desired product at any point in the amplification process by measuring fluorescence (in reality, measurement is made of its level over a given threshold). A commonly employed method of DNA quantification by quantitative PCR relies on plotting fluorescence against the number of cycles. A threshold for detection of DNA-based fluorescence is set slightly above background. The number of cycles at which the fluorescence exceeds the threshold is called the threshold cycle (Ct) or, according to the MIQE guidelines, quantification cycle (Cq).
During the exponential amplification phase, the quantity of the target DNA template (amplicon) doubles every cycle. For example, a DNA sample whose Cq precedes that of another sample by 3 cycles contained 2^3 = 8 times more template. However, the efficiency of amplification is often variable among primers and templates. Therefore, the efficiency of a primer-template combination is assessed in a titration experiment with serial dilutions of DNA template to create a standard curve of the change in Cq with each dilution. The slope of the linear regression is then used to determine the efficiency of amplification, which is 100% if a dilution of 1:2 results in a Cq difference of 1. The cycle threshold method makes several assumptions of reaction mechanism and has a reliance on data from low signal-to-noise regions of the amplification profile that can introduce substantial variance during the data analysis.
To quantify gene expression, the Cq for an RNA or DNA from the gene of interest is subtracted from the Cq of RNA/DNA from a housekeeping gene in the same sample to normalize for variation in the amount and quality of RNA between different samples. This normalization procedure is commonly called the ΔCt-method and permits comparison of expression of a gene of interest among different samples. However, for such comparison, expression of the normalizing reference gene needs to be very similar across all the samples. Choosing a reference gene fulfilling this criterion is therefore of high importance, and often challenging, because only very few genes show equal levels of expression across a range of different conditions or tissues. Although cycle threshold analysis is integrated with many commercial software systems, there are more accurate and reliable methods of analysing amplification profile data that should be considered in cases where reproducibility is a concern.
Melting curves
[edit | edit source]
qPCR permits the identification of specific, amplified DNA fragments using analysis of their melting temperature. This is usually done with double-stranded DNA-binding dyes as reporters (e.g. SYBR Green). The DNA melting temperature is specific to the amplified fragment. The results of this technique are obtained by comparing the dissociation curves of the analysed DNA samples.
Unlike conventional PCR, this method avoids the previous use of electrophoresis techniques to demonstrate the results of all the samples. This is because, despite being a kinetic technique, quantitative PCR is usually evaluated at a distinct end point. The technique therefore usually provides more rapid results and/or uses fewer reactants than electrophoresis. If subsequent electrophoresis is required it is only necessary to test those samples that real time PCR has shown to be doubtful and/or to ratify the results for samples that have tested positive for a specific determinant.
Other variants of PCR
[edit | edit source]- Assembly PCR (also known as Polymerase Cycling Assembly or PCA) is the synthesis of long DNA structures by performing PCR on a pool of long oligonucleotides with short overlapping segments, to assemble two or more pieces of DNA into one piece. During the polymerase cycles, the oligonucleotides anneal to complementary fragments and then are filled in by polymerase. Each cycle thus increases the length of various fragments randomly depending on which oligonucleotides find each other. It is critical that there is complementarity between all the fragments in some way or a final complete sequence will not be produced. After this initial construction phase, additional primers encompassing both ends are added to perform a regular PCR reaction, amplifying the target sequence away from all the shorter incomplete fragments. A gel purification can then be used to identify and isolate the complete sequence. This technique may substitute for ligation-based assembly.
- In Colony PCR, bacterial colonies are screened directly by PCR, without the need for overnight cultures and plasmid preparation. Colony PCR can be used as a high-troughput method to check whether the plasmids in the cells contain an insert. With appropriate primers, it is also possible to determine the orientation of the insert. Colonies are sampled with a sterile pipette tip and a small quantity of cells is transferred into a PCR mix. During the initial denaturation step, the plasmids are released from the cells and can serve as templates. An extended denaturation step and the use of a lysis buffer can promote cell lysis.
- Hot-start PCR is a technique performed manually by heating the reaction components to the DNA melting temperature (e.g. 95 °C) before adding the polymerase. In this way, non-specific amplification at lower temperatures is prevented. Alternatively, specialized reagents inhibit the polymerase's activity at ambient temperature, either by the binding of an antibody, or by the presence of covalently bound inhibitors that only dissociate after a high-temperature activation step. 'Hot-start/cold-finish PCR' is achieved with new hybrid polymerases that are inactive at ambient temperature and are only activated at elevated temperatures.
- Multiplex-PCR uses several pairs of primers annealing to different target sequences. This permits the simultaneous analysis of multiple targets in a single sample. For example, in testing for genetic mutations, six or more amplifications might be combined. In the standard protocol for DNA Fingerprinting, the targets assayed are often amplified in groups of 3 or 4.
- Nested PCR is used to increase the specificity of DNA amplification. Two sets of primers are used in two successive reactions. In the first PCR, one pair of primers is used to generate DNA products, which may contain products amplified from non-target areas. The products from the first PCR are then used as template in a second PCR, using one ('hemi-nesting') or two different primers whose binding sites are located (nested) within the first set, thus increasing specificity. Nested PCR is often more successful in specifically amplifying long DNA products than conventional PCR, but it requires more detailed knowledge of the sequence of the target.


- The overlap extension polymerase chain reaction (OE-PCR) is also referred to as Splicing by overlap extension/Splicing by overhang extension (SOE) PCR. It is used to insert specific mutations at specific points in a sequence or to splice smaller DNA fragments into a larger polynucleotide.
As in most PCR reactions, two primers - one for each end - are used per sequence. To splice two DNA molecules, special primers are used at the ends that are to be joined. For each molecule, the primer at the end to be joined is constructed such that it has a 5' overhang complementary to the end of the other molecule. In the first step, these elongated fragments are amplified. Afterwards, both DNA molecules are mixed and a PCR is carried out with only the primers for the far ends. The overlapping complementary sequences introduced will serve as primers and the two sequences will be fused. This method has an advantage over other gene splicing techniques in not requiring restriction sites. To get higher yields, some primers are used in excess as in asymmetric PCR.
To insert a mutation into a DNA sequence, a specific primer is designed. The primer may contain a single substitution or contain a new sequence at its 5' end. If a deletion is required, a sequence that is 5' of the deletion is added, because the 3' end of the primer must have complementarity to the template strand so that the primer can sufficiently anneal to the template DNA.
- Rapid Amplification of cDNA Ends (RACE) is used to obtain the full length sequence of an RNA transcript. RACE results in the production of a cDNA copy of the RNA sequence of interest, produced through reverse transcription, followed by PCR amplification of the cDNA copies. RACE can provide the sequence of an RNA transcript from a small known sequence within the transcript to the 5' end (5' RACE-PCR) or 3' end (3' RACE-PCR) of the RNA. This technique is sometimes called one-sided PCR or anchored PCR.
- In Touchdown PCR, the annealing temperature is gradually decreased in later cycles. The annealing temperature in the early cycles is usually 3-5 °C above the standard Tm of the primers used, while in the later cycles it is a similar amount below the Tm. The initial higher annealing temperature leads to greater specificity for primer binding, while the lower temperatures permit more efficient amplification at the end of the reaction.
References
[edit | edit source]- Bustin, S.A., 2004. A-Z of Quantitative PCR. Electrophoresis 1–15.
- Higuchi, R., Dollinger, G., Walsh, P.S., Griffith, R., 1992. Simultaneous amplification and detection of specific DNA sequences. Biotechnology. (N. Y). 10, 413–417. doi:10.1038/nbt0492-413
- Kubista, M., Andrade, J.M., Bengtsson, M., Forootan, A., Jonák, J., Lind, K., Sindelka, R., Sjöback, R., Sjögreen, B., Strömbom, L., Ståhlberg, A., Zoric, N., 2006. The real-time polymerase chain reaction. Mol. Aspects Med.
- Mullis, K.B., 1990. The unusual origin of the polymerase chain reaction., Scientific American.
- Powledge, T.M., 2004. The polymerase chain reaction. Adv. Physiol. Educ. 28, 44–50.
- Saiki, R.., Scharf, S., Faloona, F., Mullis, K.B., Horn, G.T., 1985. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science (80-. ). 239, 1350–54. doi:10.1126/science.2999980

