Methods and Concepts in the Life Sciences/Recombination
Recombination
[edit | edit source]Homologous recombination
[edit | edit source]
Homologous recombination is a type of genetic recombination in which nucleotide sequences are exchanged between two similar or identical molecules of DNA. It is most widely used by cells to accurately repair harmful breaks that occur on both strands of DNA, known as double-strand breaks. Homologous recombination also produces new combinations of DNA sequences during meiosis. Moreover, homologous recombination is used in horizontal gene transfer to exchange genetic material between different strains and species of bacteria and viruses.
Although homologous recombination varies widely among different organisms and cell types, most forms involve the same basic steps. After a double-strand break occurs, sections of DNA around the 5' ends of the break are cut away in a process called resection. In the strand invasion step that follows, an overhanging 3' end of the broken DNA molecule then "invades" a similar or identical DNA molecule that is not broken. After strand invasion, the further sequence of events may follow either of two main pathways; the DSBR (double-strand break repair) pathway or the SDSA (synthesis-dependent strand annealing) pathway. Homologous recombination that occurs during DNA repair tends to result in non-crossover products, in effect restoring the damaged DNA molecule as it existed before the double-strand break.
Homologous recombination is also used in gene targeting, a technique for introducing genetic changes into target organisms.
Red/ET
[edit | edit source]Red/ET is a commercial recombination technology based on homologous recombination. It involves a λ phage-derived protein pair, Redα/Redβ, and 50 bp homology regions. The homology regions can be chosen freely, which means that every position on a target molecule can be altered. A plasmid with an inducible promoter is used to express the recombination proteins. Because the origin of replication is temperature-sensitive, this plasmid can be conveniently removed after recombination.
Site-specific recombination
[edit | edit source]In site-specific recombination, DNA strand exchange takes place between segments possessing only a limited degree of sequence homology. Site-specific recombinases (SSRs) perform rearrangements of DNA segments by recognizing and binding to short DNA sequences, at which they cleave the DNA backbone, exchange the two DNA helices involved and rejoin the DNA strands. While in some site-specific recombination systems just a recombinase enzyme and the recombination sites is enough to perform all these reactions, in other systems a number of accessory proteins and/or accessory sites are also needed. Multiple genome modification strategies, among these recombinase-mediated cassette exchange (RMCE), an advanced approach for the targeted introduction of transcription units into predetermined genomic loci, rely on the capacities of SSRs.
Site-specific recombination systems are highly specific, fast and efficient, even when faced with complex eukaryotic genomes. They are employed in a variety of cellular processes, including bacterial genome replication, differentiation and pathogenesis, and movement of mobile genetic elements. For the same reasons, they present a potential basis for the development of genetic engineering tools.
Recombination sites are typically between 30 and 200 nucleotides in length and consist of two motifs with a partial inverted-repeat symmetry, to which the recombinase binds, and which flank a central crossover sequence at which the recombination takes place. The pairs of sites between which the recombination occurs are usually identical, but there are exceptions.
Based on amino acid sequence homology and mechanistic relatedness most site-specific recombinases are grouped into one of two families: the tyrosine recombinase family or the serine recombinase family. The names stem from the conserved nucleophilic amino acid residue that they use to attack the DNA and which becomes covalently linked to it during strand exchange.
Cre/lox
[edit | edit source]Cre/lox recombination is known as a site-specific recombinase technology, and is widely used to carry out deletions, insertions, translocations and inversions at specific sites in the DNA of cells. It is implemented both in eukaryotic and prokaryotic systems.
The system consists of a single enzyme, Cre recombinase, that recombines a pair of short target sequences called the lox sequences. This system can be implemented without inserting any extra supporting proteins or sequences. The Cre enzyme and the original Lox site called the loxP sequence are derived from bacteriophage P1.
Placing Lox sequences appropriately allows genes to be activated, repressed, or exchanged for other genes. The activity of the Cre enzyme can be controlled so that it is expressed in a particular cell type or triggered by an external stimulus like a chemical signal or a heat shock. These targeted DNA changes are useful in cell lineage tracing and when mutants are lethal if expressed globally.
The Cre/lox system is very similar in action and in usage to the FLP-FRT recombination system.
Cre recombinase
[edit | edit source]Cre recombinase (encoded by the locus originally named as "Causes recombination", with "Cyclization recombinase" being found in some references) is a tyrosine recombinase derived from the P1 Bacteriophage. The enzyme uses a topoisomerase I like mechanism to carry out site specific recombination events. It consists of 4 subunits and two domains. The total protein has 343 amino acids. Cre plays important roles in the life cycle of the P1 Bacteriophage such as cyclization of the linear genome and resolution of dimeric chromosomes that form after DNA replication.
LoxP site
[edit | edit source]LoxP (locus of X-over P1) is a site on the bacteriophage P1 consisting of 34 bp. The site includes an asymmetric 8 bp sequence, variable except for the middle two bases, in between two sets of palindromic 13 bp sequences. The exact sequence is given below; 'N' indicates bases which may vary.
Mechanism
[edit | edit source]
When cells that have loxP sites in their genome express Cre, a recombination event can occur between the loxP sites. Cre recombinase binds to the first and last 13 bp regions of a lox site forming a dimer. This dimer then binds to a dimer on another lox site to form a tetramer. Lox sites are directional and the two sites joined by the tetramer are parallel in orientation. The double stranded DNA is cut at both loxP sites by the Cre protein. The strands are then rejoined with DNA ligase in a quick and efficient process.
The products of Cre-mediated recombination at loxP sites are dependent upon the location and relative orientation of the loxP sites. DNA sequences found between two loxP sites are said to be "floxed". In this case the products of Cre mediated recombination depends upon the orientation of the loxP sites. DNA found between two loxP sites oriented in the same direction will be excised as a circular loop of DNA whilst intervening DNA between two loxP sites that are opposingly orientated will be inverted. Two separate DNA species both containing loxP sites can undergo fusion as the result of Cre mediated recombination. For example, plasmids can be joined using the variant lox sites 71 and 66.
FLP/FRT
[edit | edit source]Flp-FRT recombination is a site-directed recombination technology, increasingly used to manipulate an organism's DNA under controlled conditions in vivo. It is analogous to Cre-lox recombination but involves the recombination of sequences between short flippase recognition target (FRT) sites by the recombinase (Flp) derived from the 2µm plasmid of Saccharomyces cerevisiae.
The 34bp minimal FRT site sequence has the sequence
5'GAAGTTCCTATTCtctagaaaGtATAGGAACTTC3'
for which flippase (Flp) binds to both 13-bp 5'-GAAGTTCCTATTC-3' arms flanking the 8 bp spacer, i.e. the site-specific recombination (region of crossover) in reverse orientation. FRT-mediated cleavage occurs just ahead from the asymmetric 8bp core region (5'tctagaaa3') on the top strand and behind this sequence on the bottom strand. Several variant FRT sites exist, but recombination can usually occur only between two identical FRTs but generally not among non-identical ("heterospecific") FRTs.
Many available constructs include an additional arm sequences (5'-GAAGTTCCTATTCC-3') one base pair away from the upstream element and in the same orientation:
5'GAAGTTCCTATTCcGAAGTTCCTATTCtctagaaaGtATAGGAACTTC3'
This segment is dispensable for excision but essential for integration, including Recombinase-mediated cassette exchange.
Because the recombination activity can be targeted to a selected organ, or a low level of recombination activity can be used to consistently alter the DNA of only a subset of cells, Flp-FRT can be used to construct genetic mosaics in multicellular organisms. Using this technology, the loss or alteration of a gene can be studied in a given target organ of interest, even in cases where experimental animals would not survive the loss of this gene in other organs (spatial control). The effect of altering a gene can also be studied over time, by using an inducible promoter to trigger the recombination activity late in development (temporal control) - this prevents the alteration from affecting overall development of an organ, and allows single cells lacking the gene to be compared to normal neighboring cells in the same environment.
Applications
[edit | edit source]Gene knockout
[edit | edit source]
A gene knockout (abbreviation: KO) is a genetic technique in which one of an organism's genes are made inoperative ("knocked out" of the organism). Also known as knockout organisms or simply knockouts, they are used in learning about a gene that has been sequenced, but which has an unknown or incompletely known function. Researchers draw inferences from the difference between the knockout organism and normal individuals.
Knockout is accomplished through a combination of techniques, beginning in the test tube with a plasmid, a bacterial artificial chromosome or a different DNA construct, and proceeding to cell culture. Individual cells are genetically transfected with the DNA construct. Often the goal is to create a transgenic animal that has the altered gene. If so, embryonic stem cells are genetically transformed and inserted into early embryos. Resulting animals with the genetic change in their germ-line cells can then often pass the gene knockout to future generations.
The construct is engineered to recombine with the target gene, which is accomplished by incorporating sequences from the gene itself into the construct. Recombination then occurs in the region of that sequence within the gene, resulting in the insertion of a foreign sequence to disrupt the gene. With its sequence interrupted, the altered gene in most cases will be translated into a nonfunctional protein, if it is translated at all.
Because the desired type of DNA recombination is a rare event in the case of most cells and most constructs, the foreign sequence chosen for insertion usually includes a reporter. This enables easy selection of cells or individuals in which knockout was successful. Sometimes the DNA construct inserts into a chromosome without the desired homologous recombination with the target gene. To eliminate such cells, the DNA construct often contains a second region of DNA that allows such cells to be identified and discarded. Such a system is illustrated in the figure below.

In diploid organisms, which contain two alleles for most genes, and may as well contain several related genes that collaborate in the same role, additional rounds of transformation and selection are performed until every targeted gene is knocked out. Selective breeding may be required to produce homozygous knockout animals.
Conditional knockout
[edit | edit source]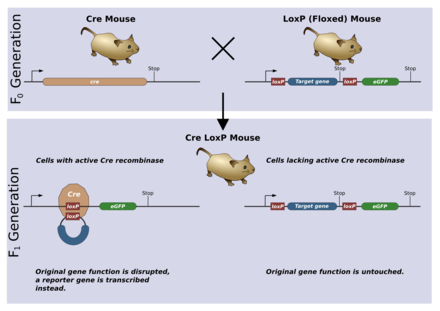
A conditional knockout allows gene deletion in a tissue or time specific manner. This is most often achieved using the Cre/lox system. The loxP sequences will be introduced into the germ-line via the same mechanism as for a normal knockout. This germ-line can then be crossed to another germ-line containing the gene for Cre recombinase. Typically, the gene will be controlled by a promoter which is tissue specific or only active during a distinct time in ontogeny. When Cre is expressed, it recombines the loxP sites and deletes the floxed gene.
Knockouts are primarily used to understand the role of a specific gene or DNA region by comparing the knockout organism to a wildtype with a similar genetic background.
Knockouts organisms are also used as screening tools in the development of drugs, to target specific biological processes or deficiencies by using a specific knockout, or to understand the mechanism of action of a drug by using a library of knockout organisms spanning the entire genome, such as in Saccharomyces cerevisiae.
Knock-in is similar to knock-out, but instead it replaces a gene with another instead of deleting it.
Procedure of producing knockout mice
[edit | edit source]There are several variations to the procedure of producing knockout mice; the following is a typical example.


- The gene to be knocked out is isolated from a mouse gene library. Then a new DNA sequence is engineered which is very similar to the original gene and its immediate neighbour sequence, except that it is changed sufficiently to make the gene inoperable. Usually, the new sequence is also given a marker gene, a gene that normal mice don't have and that confers resistance to a certain toxic agent or that produces an observable change (e.g. colour or fluorescence).
- Stem cells are isolated from a mouse blastocyst (a very young embryo) and grown in vitro. For this example, we will take stem cells from a white mouse.
- The new sequence from step 1 is introduced into the stem cells from step 2 by electroporation. By the natural process of homologous recombination some electroporated stem cells will incorporate the new sequence with the knocked-out gene into their chromosomes in place of the original gene. The chances of a successful recombination event are relatively low, so the majority of altered cells will have the new sequence in only one of the two relevant chromosomes - they are said to be heterozygous.
- The stem cells that incorporated the knocked-out gene are isolated from the unaltered cells using the marker gene from step 1. For example, the unaltered cells can be killed using a toxic agent to which the altered cells are resistant.
- The knocked-out stem cells from step 4 are inserted into a mouse blastocyst. For this example, we use blastocysts from a grey mouse. The blastocysts now contain two types of stem cells: the original ones (from the grey mouse), and the knocked-out cells (from the white mouse). These blastocysts are then implanted into the uterus of female mice, where they develop. The newborn mice will therefore be chimeras: some parts of their bodies result from the original stem cells, other parts from the knocked-out stem cells. Their fur will show patches of white and grey, with white patches derived from the knocked-out stem cells and grey patches from the recipient blastocyst.
- Some of the newborn chimera mice will have gonads derived from knocked-out stem cells, and will therefore produce eggs or sperm containing the knocked-out gene. When these chimera mice are crossbred with others of the wild type, some of their offspring will have one copy of the knocked-out gene in all their cells. These mice will be entirely white and are not chimeras, however they are still heterozygous.
- When these heterozygous offspring are interbred, some of their offspring will inherit the knocked-out gene from both parents; they carry no functional copy of the original unaltered gene (i.e. they are homozygous for that allele).
