Structural Biochemistry/Cell Signaling Pathways/Insulin Signaling
Definition
[edit | edit source]Insulin is a hormone released by pancreatic beta cells in response to elevated levels of nutrients in the blood. Insulin causes cells in the liver, muscle, and fat tissue to take glucose from the blood, promoting the storage of these nutrients as glycogen in the liver and muscle and stop using fat as an energy source. When fails to control the insulin levels, diabetes will result. Patients with Type 1 diabetes is characterized by the inability to produce the hormone internally, whereas in Type 2 diabetes, the body becomes resistant to the effects of insulin presumably due to the failure of controlling glucose levels. As a peptide hormone, insulin consists of 51 amino acids and has a molecular weight of 5808Da. Produced in the islets of Langerhans in the pancreas, insulin's name stems from Latin insula for "island."
Insulin Receptor Structure
[edit | edit source]
The receptor of insulin is a dimer of two identical subunits that spans the cell membrane. Each of the two subunits is made of one α-chain and one β-chain, connected together by a single disulfide bond. The α-chain lies on the exterior of the cell membrane, while the β-chain spans the cell membrane in a single segment, and with the exception of this segment lies on the inside of the cell membrane.[1]
Activation of Insulin Receptor
[edit | edit source]The two α-chains on the exterior of a cell move together when insulin is detected and fold around the insulin. This action moves the β-chains together, thus making the β-chains an active tyrosine kinase. The tyrosine kinase catalyzes the transfer of phosphoryl groups from ATP to tyrosine in the activation loops of the β-chains. The phosphorylated activation loop then drastically changes conformation, causing the kinase to become fully active.[2]
Insulin-Receptor Substrates (IRS)
[edit | edit source]Insulin-Receptor Substrates are a special group of proteins that are attracted to the phosphorylated sites on the activated Insulin Receptor. These sites act as a docking point for IRS proteins. Each IRS molecule has four sequences that are approximately Tyr-X-X-Met. This reoccurring sequence is responsible for the IRS molecules' affinity of the receptor tyrosine kinase. The tyrosine kinase then phosphorylates these Tyrosines, causing the IRS molecules to activate. Activated IRS proteins act as adaptor proteins. Adaptor proteins bring a kinase and its substrate together rather than activating a kinase. An example would be an activated IRS protein binding to a lipid kinase, thus attracting the lipid kinase to lipid membrane, its substrate.[3]
Example of a specific IRS pathway
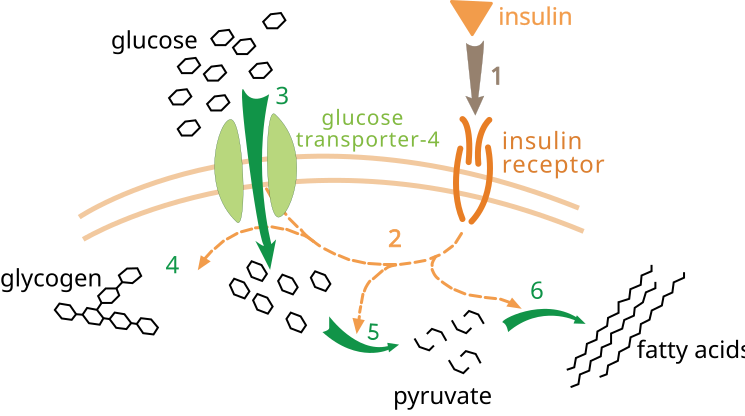
[edit | edit source]SH2 is a domain present in many signal-transduction proteins. SH2 domains are host to specific phosphotyrosine sequences, such as those in activated IRS proteins. This is a specific process, so each SH2 domain has a binding preference to an approximate sequence including phosphotyrosine. A large group of lipid kinases have SH2 domains that are attracted to IRS proteins. These lipid kinases react at the 3-position of inositol in phosphatidylinositol 4,5-bisphosphate (PIP2) and add a phosphoryl group. By attracting the lipid kinases to IRS proteins, the reaction moves the kinases to PIP2 located on the membrane where it can phosphorylate them to phosphatidyl-inositol 3,4,5-trisphosphate (PIP3) This causes a cascade which activates the protein kinase PDK1, which phosphorylates the protein kinase Akt, thus activating it as well. Akt is a free moving kinase inside the to cytoplasm, and moves around phosphorylated targets within the cell, such as proteins that control the movement GLUT4, a glucose transporter, bringing it to the cell surface. This is only one of many pathways that begin with the activation of IRS by an insulin receptor.[4]
Mechanism of Insulin Action
[edit | edit source]The phosphorylation of the insulin receptor starts up serine-threonine phosphorylation of a series of proteins. They are coupled to 3 additional protein kinase signal systems. The protein kinase signal systems include: 1) The pathways signaling through PI3-kinase and phosphatidylinositol (3,4,5)P3 (PI-3 kinase and protein kinase B/Akt), 2) Mitogen-activated protein kinases (MAPKinases), 3) Possible interaction via kinases not coupled to IRS proteins.
Insulin Pathway
[edit | edit source]•Glucose storage and uptake
Insulin binds to a-subunit and changes the conformation causing autophosphorylation of tyrosine residues. Those residues are then picked up by phosphotyrosine-binding domains known as PTB, such as insulin receptor substrate, SHC and Cbl. Activated receptors phosphorylate tyrosine residues on the receptors. These receptors interact by signaling molecules through their SH2 domains and activates many other pathways such as PI 3-kinase signaling, MAPK activation and Cbl/CAP complex activation. Regulation of glucose, lipids and protein metabolism are the results of these pathways.
•Protein synthesis
Insulin stimulates the uptake of amino acids into the cell, stops protein degradation and promotes protein synthesis.
•Regulation of lipid synthesis
Insulin stimulate the uptake of fatty acids and lipid synthesis, as well as inhibiting lipolysis. Lipid synthesis requires a lot of transcription factor steroid regulatory element-binding proteins (SERBP)-1c. Insulin decrease the concentration of cAMP through the activation a cAMP specific phosphodiesterase in adipocytes to inhibit lipolysis, lipid metabolism.
•Mitogenic responses
Another substrate for the insulin receptor is SHC. The phosphorylation of SHC is related to GRB2 which can activate the MAPK pathway independently of an insulin recptor substrate. Other signal transduction proteins interact with GRB2, an insulin receptor substrate that contain the SH3 domains which is related to the guanine nucleotide exchange factor son-of sevenless and promotes the activation of the MAPK pathway, then reaches mitogenic responses.
References
[edit | edit source]- ↑ Berg, Jeremy (2007). Biochemistry, 6th Edition. New York, New York: Sara Tenney. pp. 392–393. ISBN 978-0-7167-8724-2.
{{cite book}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ↑ Berg, Jeremy (2007). Biochemistry, 6th Edition. New York, New York: Sara Tenney. pp. 392–393. ISBN 978-0-7167-8724-2.
{{cite book}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ↑ Berg, Jeremy (2007). Biochemistry, 6th Edition. New York, New York: Sara Tenney. pp. 393–394. ISBN 978-0-7167-8724-2.
{{cite book}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ↑ Berg, Jeremy (2007). Biochemistry, 6th Edition. New York, New York: Sara Tenney. pp. 394–395. ISBN 978-0-7167-8724-2.
{{cite book}}: Unknown parameter|coauthors=ignored (|author=suggested) (help)
http://www.abcam.com/index.html?pageconfig=resource&rid=10602&pid=7