Structural Biochemistry/DNA recombinant techniques/History and Study of Bacteriophage Lambda
Overview
[edit | edit source]Its linear genome was observed to form a circle via complementary base pairing when entering a host cell. The genome was able to act in such a peculiar manner due to the presence of single stranded areas called cohesive sites (cos). These cos sites are able to base pair, and the nature of these sites are the same as those produced by restriction endonucleases. (Ex: EcoR1 produces "sticky ends"). This circular phage DNA is able to recombine with the host cell DNA via attachment sites. Progeny of the phage with the addition of some bacterial gene take place by the lysogenic pathway. What makes lambda phage extremely advantageous is the fact that it can destroy its host (lytic pathway) or it can become part of its host (lysogenic pathway). Studies on the control of these alternative cycles have been very important for our understanding of the regulation of gene transcription.
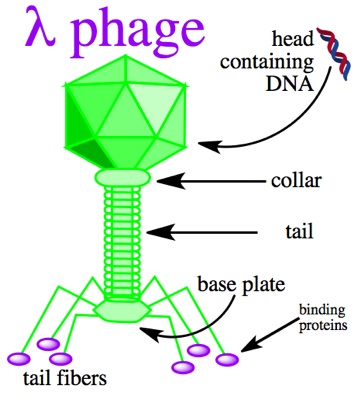
Lambda phage is a virus particle consisting of a head made up of double-stranded linear DNA as its genetic material, and a tail. It infects the host cell by injecting its own DNA through the tail at which point the phage will enter the lytic or lysogenic pathway. Large segments of the 48 kilobase pair DNA of the lambda phage are not essential for productive infection and can be replaced by foreign DNA, thus making lambda phage an ideal vector.
History
[edit | edit source]Lambda phage was first isolated by Esther Lederberg in 1950 from Escherichia coli. It has been an intensely studied organism, and has been a useful tool in molecular biology. Lambda phage can be used for cloning of recombinant DNA, the use of its site specific recombinase (int) for the shuffling of cloned DNAs by the 'Gateway' method, and in recombineering. The discovery and study of lambda phage led to an understanding of transduction which provides a major mechanism that can explain modes of evolution.
Bacteriophage Lambda was studied by Allan Campbell in 1962 who studied the phage's integration process. He observed that lambda phage had a unique characteristic what some phages would infect and reproduce in some strains of E. coli while other strains seemed immune. Probing further into why this occurred lead to the discovery that the immune strains contained a dormant copy of the phage, but the dormant copy could be activated. Study the phage provided priceless contributions into virus life cycles, the regulation and expression of genetic material, and the mechanism of integration and excision of genetic material.
Mutant Lambda Phages
[edit | edit source]Mutant phages have been genetically designed to make cloning easier. And especially useful one is called lambda(gt)-(lambda)(beta) which contains only two EcoRI cleavage sites instead of the normal five. After cleavage, the middle segment of this phage DNA can be removed leaving 72% of the normal genome length which is too little to be packaged into a lambda particle. Thus, a suitably long DNA insert between the lambda segments of DNA enables a recombinant DNA molecules to be packaged. These modified viruses enter bacteria much more easily than plasmids.
Genomic Library
[edit | edit source]A genomic library is a large collection of DNA fragments. Such libraries assist in identifying DNA fragments that have a gene of interest. Creating a genomic library utilizes lambda phage. First, total genomic DNA is mechanically sheared to produce a random population of overlapping DNA fragments which are separated by gel electrophoresis. All fragments that are about 15 kilobase pairs long are isolated and attached with synthetic linkers to insert into a lambda phage which replicated themselves and then lyse their bacterial hosts. The resulting lysate contains fragments of DNA houssed[check spelling] in virus particles which constitute a genomic library.
Restriction & Modification Systems
[edit | edit source]Restriction: Phages grown in one bacterial cell fail to grow in other bacterial strains. The degradation of phage lambda DNA was observed in certain bacterial strains. This was later found to be a result of restriction endonucleases which would cut the phage DNA after detecting it as being foreign. Thus, restriction endonucleases was termed because of its ability to restrict foreign DNA.
Modification: Phages are modified such that they can be grown normally in other bacterial strains. An example of modification is the methylation of lambda phage DNA so that it would not be cut by restriction endonuclease.
References
[edit | edit source]Allison, et al. Fundamental Molecular Biology, 2007.