Structural Biochemistry/Gatekeepers of Chromatin: Small Metabolites Elicit Big Changes in Gene Expression
Introduction
[edit | edit source]Flexible chromatin is needed for eukaryotes to constantly tuning their gene expression in transcript levels in order for them to respond to the environment. Such constant rapid changes in gene expressions can be done through post-translational modifications of histone proteins, which controls the structure of chromatin. Recent studies have revealed that several particular metabolites, in fact, may have shown to be the key regulators that link gene expression with cellular metabolism.
Chromatin Architecture and Histone Modifications
[edit | edit source]In eukaryotes, 2 meter of 146bp DNA is condensed and packaged into chromatin structure by wrapping it around an octamer, which contains two copies of each histone proteins H2A, H2B, H3 and H4, in order to fit inside a nucleus and at the same time, allow access to the genetic material for replication, repair and transcription in such compacted form. The amino terminal of every lysine-riched histones are exposed to modifications such as acetylation, methylation, phosphorylation, ubiquitylation, SUMOylation and poly-ADP-eibosylation with the help of modifying enzymes, which uses metabolites such as ATP, NAD+, actyl-coenzyme A and S-adenosylmethionine.

Link Between Chromatin and Metabolism
[edit | edit source]The first study of chromatin-level gene regulation was done by Allfrey, in 1964. Allfrey hypothesized that RNA synthesis is closely related to histone acetylation. It was thought that the positively charged lysine tail of histone and the negatively charged backbone are neutralized through acetylation for transcription activities. There are many transcription factors that has bromodomain, which interacts specifically with lysine after acetylation. In addition, acetylation is also important for binding trans-acting factors and chromatin remodeler. Support of this hypothesis was actually first discovered when studying the “silent” chromatin in Saccharomyces cerevisiae. The silent information regulator, a subset of histone deacetylases that is also known as SIR/sirtuin, was found to be responsible for keeping the amino group of histone H3 and H4 in specific genome region in hypoacetylated form. Other sirtuins, such as yeast sir2, were also found to decetylate lysine residue on histone by NAD+, which is used as substrate. The fact that sirtuins are regulated by NAD+ levels suggests a strong connection between metabolism and gene expression regulation.
Acetyl-CoA and Histone Acetylation
[edit | edit source]Histone acetylation was first discovered and identified by Brownell and Allies in the mid 1990s when studying polypeptide in Tetrahymena Thermophila. This study revealed that there is histone acetyltransferase (HAT) activity within the polypeptide chain, which is similar to yeast Gcn5 transciptional coactivator, thus showing a direct relationship between protein and histone acetylation that affects gene transcription. Besides HAT, lysine acetyltransferase (KAT) activity had also been identified to be responsible for acetylation. Both HAT and KAT were found to use acetyl-coenzyme A (acetyl-CoA), an important metabolite that accounts for many metabolic reactions in a cell, as acetyl donor during the process of acetylation. This shows that acetyl-CoA production may be crucial in acetyltransferase regulation and that the levels of acetyl-coA may affect or limit the modifications of histone. In yeast, enzyme that synthesize acetyl-CoA , acetyl-CoA synthetases Acs1p and Acs2p, are shown to be the key regulators of chromatin and gene expression. Studies have proven that mutation of either enzyme leads to growth defects and loss of H3/H4 acetylation, thus altering gene transcription. Similar cases occur in mammalian acetyl-CoA-producing enzymes such as ACL. Further studies have found that acetyl-CoA is not only important for histone acetylation, but also important for modification of nonhistone acetylation. Many cellular processes such as DNA repair, cell cycle progression, differentiation, replication, and apoptosis are found to be regulated by the acetylation of nonhistone proteins.
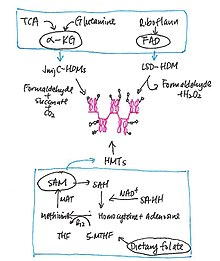
SAM and Histone Methylation
[edit | edit source]S-adenosylmethionine (SAM), which is produced by SAM synthetases called methionine adenosyl transferase (MAT), gives off its methyl groups to methyltransferases, HMTs, to carry out DNA, RNA and proteins methylation, as well as nonhistone methylation. As shown in figure “Methylation and demethylation of histone”, SAM can also be converted to S-adenosylhomocystein (SAH), which can act as inhibitor of methyltransferases if its level is high.
Through recent studies, SAM has shown a close connection with gene repression. When there is a single point mutation, the synthesis of SAM is prevented due to the breakdown of MatIIα, thus the transcriptional repression is reversed. Moreover, the demethylation of H3K4 and H3K9 greatly decreases.
α-Ketoglutarate (KG) and Histone Demethylation
[edit | edit source]For demethylation, histone demethylases (HDMs) act to remove the methyl groups from histone. As shown in figure "Methylation and Demethylation of Histone",α-Ketoglutarate (α-KG), which is also known as 2-oxoglutarate), is utilized as a substrate by Jmjv-domain-continaing histone demethylases (HDMs) to demethylate histones, while flavin adenine dinucleotide (FAD) is used as cofactor by lysine-specific histone demethylases (LSD). When mutation occurs in isocitrate dehydrogenase (IDH) genes, enzymes are not able to convert isocitrate to α-KG during the TCA cycle, but instead, a competitive inhibitor of HDM called 2-hydroxyglutarate (HG) are produced, which would lead to transcriptional repression. Furthermore, another α-KG-dependent enzyme called TET2 would also greatly reduce. All these would affect the methylation of DNA and thus alter the gene expression.
Reference
[edit | edit source]Kaochar, Salma and Benjamin P. Tu. “Gatekeepers of chromatin: Small metabolites elicit big changes in gene expression.” Trends in Biochemical Sciences. 37.11 (2012):477-483. <http://www.sciencedirect.com/science/article/pii/S0968000412001120>.
