Structural Biochemistry/Gene Flow
Gene flow is the movement of genes from one population to another through the transfer of alleles. This can occur within a population as well as across a population. Gene flow includes lots of different kinds of events, such as pollen being blown to a new destination or people moving to new cities or countries. If genes are carried to a population where those genes previously did not exist, gene flow can be a very important source of genetic variation. In the graphic below, the gene for brown coloration moves from one population to another.
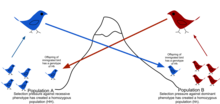
The rate of gene flow depends on the capability of individual organisms. For example, a wind-pollinated corn will have a possible gene flow range of about 50 feet whereas fruit flies have migrated from over 15km.
Random Drifts
[edit | edit source]Random drifts: the idea of allele frequencies in gene flow is that all alleles have an equal opportunity of being transfered. Although some alleles appear to be more favorable, this is accounted by its increased appearance in the population. Random drift is the important mechanism of evolution. One aspect of genetic drift is the random nature of transmitting alleles from one generation to the next, given that only a fraction of all possible zygotes become mature adults. The easiest case to visualize is the one which involves binomial sampling error. If a pair of diploid sexually reproducing parents (such as humans) have only a small number of offspring, then not all of the parent's alleles will be passed on to their progeny due to chance assortment of chromosomes at meiosis. In a large population, this will not have much effect in each generation because the random nature of the process will tend to average out. But in a small population the effect could be rapid and significant.
Gene Flow in Humans
[edit | edit source]Gene flow in humans can be primarily accounted by migration. For example, the Vietnam War caused the mating of US soldiers with Vietnamese women, thus leading to a differing gene pool frequency. The loss or gain of people can also affect the gene flow. For example, if red-haired Scottish were to move out of the country, then the next generations of Scottish would consist of fewer red-heads. Population genetic studies are mainly based on the description of genetic variability and on interpopulational comparisons using genetic distance measures. The evolutionary dynamics of the populations are inferred from these parameters and accurate estimates of gene flow may be critical. The present study reevaluates the role of gene flow in human populations by different statistical methods from a number of microsatellite and protein polymorphism data. The estimated number of individuals exchanged per generation (Nm) was greater than 1 in all data sets with all statistical methods. The correlation between geographic and genetic distances suggests a pattern of isolation by distance, characteristic of demographic and genetic equilibrium conditions among populations worldwide. Thus the high values of Nm may be interpreted as a reflection of high gene flow between geographically close populations. As expected, gene flow appears to exert a pivotal role in the genetic history of humans.
Gene Flow between Species
[edit | edit source]Gene flow can occur between species through viruses. The DNA of one cell is extracted by a virus and injected into another. The extent of this type of gene flow between humans is unclear at the moment, although estimates of 40-50% are plausible. However, this can be observed in several species such as insects, fish, reptiles, mammals, and especially microorganisms.
Researchers hypothesize that differential gene flow across species boundaries is a pattern resulting from selection, in which the absence of selection on some traits or alleles between species results in gradual transitions of these traits across a hybrid zone. In contrast, ongoing selection to maintain species at other traits prevents the dispersal of alleles across species boundaries and is expected to produce a pattern of steep transitions of traits across a hybrid zone. In house mice, researchers found that alleles near the center of the X chromosome exhibit steep transitions across the hybrid zone, and these alleles are in a region previously shown to be associated with hybrid sterility. This work demonstrates the utility of hybrid zones to test for the effect of selection on interspecies gene flow in particular regions of the genome.
By continuing to study hybridizing species and hybrid zones, researchers can further understand which sets of genes have permissible gene flow and which are more restricted. Then, by studying genes in the latter category, scientists can learn which genes or gene sets may be important in maintaining species integrity in the face of gene flow.
Allele Frequency in Subpopulations
[edit | edit source]The average allele frequency can be calculated via this formula: pt = p + (po - p ) (1-m)^t
p is the frequency of alleles in generation t m is the migration rate o is the initial frequency of an allele in a subpopulation t is the number of generations
OMPG is used to provide a balance between drift and gene flow and allow heterozygosity by minimizing the loss of alleles.
Given subpopulation size, number of subpopulations, the migration rate m, generations to evolve, this script shows changes of allele frequencies among subpopulations at single locus, which results from the effect of migration. All subpopulations have equal number of individuals. Migration is depicted in the form of a N*N matrix, which is consisted with identical non-zero values at non-diagonal positions and zero on the diagonal line, where N is the number of subpopulations. Sum of values of elements on row i (i = 1, 2, …, N) of the migration matrix equals to the migration rate, thus it ensures that every individual in subpopulation i has probability m to become a migrant, and such a migrant has equal possibility of entering any other subpopulation. Initialization of allele frequencies among subpopulations is to use a list of numbers, which have counts equal to number of subpopulations and been set evenly distributed between 0 and 1, including 0 and 1. Therefore, the average allele frequency named theoretical value that all subpopulations share after a long term of evolution will be 0.5.
Reference
[edit | edit source]- http://evolution.berkeley.edu/evosite/evo101/IIIC4Geneflow.shtml
- Gene flow at Wikipedia
- Gene flow at Encyclopædia Britannica
- www.life.illinois.edu%2Fib%2F443%2Flectures%2FEVOLGEN4.PPT
