Structural Biochemistry/Synthetic Biology
Synthetic Biology, An Overview
Synthetic biology is an emerging field by which scientists combine principles of biology and engineering in order to produce biological functions in organisms that may or may not otherwise occur naturally. One of the aims of synthetic biology is to understand the many interactions in living cells and by fabricating biological systems and understanding how they function. Since natural biological systems are so complex, scientists in this field start by making simple synthetic systems and then studying what factors affect that fabricated system. In this way, the "design" of future synthetic systems can be continually improved as well as gaining a deeper insight to the complex interactions within those biological systems. Thus, the idea is to understand the complex interactions in living systems by building and designing them from bottom to top. Originally, this was the aim of the field of systems biology, which aims to understand the complexity of living systems by taking all the biological interactions as a whole and then putting forth models in order to describe how they give rise to intricate functions. Synthetic biology has the same goal, except these interactions are studied by the attempt to reproduce them. Future advances in this field also have a great potential to discover new drugs and methods by which to cure disease.

One of the ways by which synthetic biologists construct and study fabricated biological systems is by constructing genetic oscillators. These oscillators are seen as "circuits" which can be designed in order to control gene expression in a cell. Such oscillators are designed in order to further investigate the natural biological rhythms that occur in living entities. Such rhythms can be affected by external factors, such as day and night, and affect physiological behavior such as sleep and migrating patterns. In order to understand how these genetic oscillators work, it is important to understand the basics of molecular biology. The "Central Dogma of Molecular Biology" states that proteins are produced by DNA which is transcribed into mRNA (messenger RNA), which is later "translated" to produce the protein. The protein is coded for from a specific gene. The transcription of this gene is controlled by a promoter, called RNA polymerase, whose ability to catalyze the transcription is controlled by proteins that can either hinder its progress or catalyzing its progress. The former proteins are termed "repressors", and the latter proteins are termed "activators". These proteins act between the DNA and the RNA polymerase in order to perform their functions. Although this process is not true for all genes, as some are always needed to be expressed, it is important for understanding how the other genes are expressed when the need for them arises. An example of a synthetic genetic oscillator may be when there are a system of several genes in a plasmid (circular region of DNA) in which each of the genes encode for proteins that act as repressors for one of the promoters of one of the other genes in the system. In this way, once one of the genes is "activated", it produces proteins which represses the transcription of one of the other genes in the system, which in turn represses another gene which represses the first. Such a system can lead to oscillations in gene expression,termed a negative feedback loop, which can be observed if one of the genes encodes for a fluorescent protein. These oscillations can be mathematically modeled and analyzed much in the same way electronic control systems are analyzed. Advances in this field can lead to genetic oscillators that can be designed to fit control parameters that describe their behavior.
The synthetic genetic oscillators are built using principles of recombinant DNA technology. Genes and their promoters to be included in the recombinant plasmid are cut and amplified by PCR. The original plasmid is cut by enzymes and the DNA is then ligated to the original plasmid. This procedure is repeated for the rest of the genes to be included in the oscillator. They are later transformed into cells in order to study the effects of the oscillator.
Defining Synthetic Biology Space
Synthetic biology can exist in different levels of complexity and divergence from nature depending on the complexity of biological units involved and the specific field employed to study these biological components. Recently, Derek Woolfson et al. proposed the idea of synthetic biology space in order to organize the different complexities of biological componenets involved and the different approaches taken to test the components in the context of synthetic biology.[1]
The different biological components are divided in order of complexity: basic units, tectons, self assembled units, and functional assemblies. Basic units are defined as amino acids, nucleic acids, sugars, and lipids. In addition to these building blocks, synthetic biology aims to increase the number of building blocks that synthetic biologists can work with. It has even been shown that these building blocks can be made that completely diverge from their natural counterparts. However, Woolfson et al. show that these synthetic building blocks have two major drawbacks: it is not known whether these synthetic basic units can work in living systems, and there is no template for their replication in living systems. Tectons are the next level up the chain of biological components in terms of complexity, they refer to a chain of basic units that have yet to assemble to higher order structures. Examples of tectons could include alpha helicies and beta strands in proteins. Synthetic tectons, specifically DNA, having a variety of two and three dimensional forms, are possible to be made. It much less understood when it comes to assembling synthetic tectons from peptides. However, advances have been made in the case of coil coiled alpha helicies, as they contain information to further assemble into different conformations.
-
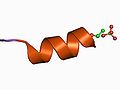
An example of a basic unit, an amino acid.
-
The basic units form a tecton, here shown as an alpha helix.
-
The tectons form self-assembled units, shown here as a protein composed of alpha helicies.
Self-assembling units have enough information to self-assemble into a discrete unit. Advances have been made in synthetically designing self-assembling units by using computer aided programs such as Rosetta Design. The program works by taking into account the amino acid sequence that would give the least sterically hindered (most energetically favorable) in a self-assembled unit. An alternative method is used by keeping a database of natural peptide strands in proteins and merging them to form a more complex peptide. It has also been shown that proteins with "switching sequences" can designed. These refer to proteins that have different conformational folds depending on their environment. The most complex biological components used for synthetic biology are termed functional assemblies. These functional assemblies can be fused together by synthetic biologists to make new components with a variety of different shapes and geometries.
As mentioned earlier, there are four different approaches that can be taken for synthetic biology depending on the complexity of biological components used and their divergence from the natural world. They are genome engineering, biomolecular engineering, biomolecular-design, and proto-cell design. Genome engineering deals primarily with manipulating and constructing DNA sequences. Biomolecular engineering uses the ability to fuse different functional assemblies in order to produce certain functionalities. biomolecular-design deals with biological design with the biomolecular components. And, seemingly far-fetched proto-cell design aims to produce an entirely self-sustained biological system from synthetic parts. One of the major challenges to this end is to produce synthetic encapsulation material in the form of membranes that has suitable fluidity and permeability. Another challenge involves the possible design of integral membrane proteins.
References
[edit | edit source]- ↑ K. Channon, E. H. C. Bromley, D. N. Woolfson, Curr. Opin. Struct. Biol. 2008, 18, 491– 498.