Structural Biochemistry/p53
p53 is one of the most extensively studied tumor suppressors in multicellular organisms. It was discovered in 1979 and over 60,000 papers have been published about it.[1] It is nicknamed "Guardian of the Genome" and in 1993 it was named the "Molecule of the Year" by Science Magazine.[1] It is also known as “protein 53” or “tumor protein 53” and is part of the small p53 family which also includes p63 and p73[2]. This tumor suppressor is regulated by a gene called “TP53 gene”. p53 works by sensing oncogenic cytotoxic and genotoxic stress signals and responds by signaling for cell-cycle arrest as well as apoptosis in order to prevent the cells from further growing. Several experiments have shown that loss of p53, due to inactivation, rearrangement or some other change that causes the protein to lose it's function, promotes the growth and survival of cancer. [3]. It is estimated that 50% of the people who have cancer have a mutation in the p53 protein [1]
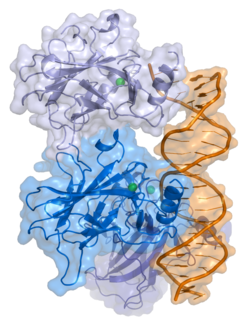
The tumor suppressor p53 in humans is a polypeptide of 393 amino acids that take up five domains. p53 is active as a tetramer that has four identical chains of 393 residues. The N-terminal region consists of disordered transcription domains and a proline rich region. At the C terminus, p53 contains the regulatory domain of unfolded basic amino acids that binds DNA nonspecifically as a transcription factor. [4] The protein is normally found in the cytoplasm of the cell. If the cell is properly functioning and the p53 protein is not needed, then it is degraded by the MDM2 protein.[1] If the cell is stressed or there is something wrong, then p53 is passed to the nucleus where is dimerizes and eventually tetramerizes to act as a transcription factor.[1].
Structure
[edit | edit source]p53 is a protein which regulates the cell cycle. There are three domains in the p53 protein that are responsible for directly DNA repair or cell death.[1] The three domains are responsible for binding to DNA, recruiting proteins (other transcription factors) to the DNA, and the third domain is a regulatory domain in the C-terminus that regulates the binding of p53 to DNA. [1] The wild-type p53 is a labile protein that is comprised of folded and unstructured regions which function in a synergistic manner.[1]
N Terminus p53
[edit | edit source]The N terminus of p53 has unfolded regions, but has secondary residual structures that contain hydrophobic resides. The acidic TAD, the main body, which has 2 ill-defined subdomains (TAD1 & TAD2), has binding sites for multiple interactions with proteins that deal with transcription machinery and transcriptionary coactivators. Thanks to the intrinsic disorder of TAD it can facilitate the binding of diverse proteins with high specificity. The n terminus of p53 deals with post translation modification where multiple phosphorylation of serine and threonine residues, by several protein kinases, happen which shift the affinity for different proteins that compete to bind with p53. The proline rich region that is in the n terminus links the TAD to the DNA binding domain of the humans. The PXXP motifs, which are in p53 help modify and mediate the protein to protein interactions. [4]
DNA-Binding Domain
[edit | edit source]The DNA binding site for p53 is 20 nucleotides long. As the protein is a tetramer, this means that each dimer binds to 10-basepair sequences that have the general pattern Pu Pu Pu C A/T A/T G Py Py Py from the 5' to 3'direction.[1] Thanks to the crystal and solution structure of p53C, the structures have been found in which there are complexes within the p53C that bound to domains of signaling protein. This consists of immunoglobulin beta sandwich that is able to provide a scaffold for DNA-binding surfaces. There is a loopsheet helix motif which dock to the DNA major grooves including loop L1, and beta strands of S2 & S2'. Although these loops are large, they are stabilized by a zinc ion. If this zinc is lost, there will a be a decrease in the thermodynamic stability and a increase in the aggregation tendencies. [4]
C Terminus
[edit | edit source]The C terminus consists of regions in the protein that help tetramerization. With the help of 3 residues, it forms the central hydrophobic core of the dimer. Formation of stable dimers are through high protein concentrations in the solution. Some extreme C terminus may be intrinsically disordered through local disorders to order transitions from binding to nonspecific DNA or proteins. Some motifs from residues may affect the confirmation. The C terminus is required for efficient binding and transactivation of target genes in large molecules of DNA. [4]
Mechanism p53 plays an important role in cell cycle control and apoptosis. The defective p53 could also allow abnormal cells to proliferate, which then results in cancer. When the DNA is damaged, it will trigger the increase of p53 proteins, which have three major functions. They are DNA repair, growth arrest, and apoptosis (cell death). The cellular concentration of p53 must be tightly regulated. However, the high level of p53 may accelerlate the aging process by excessive apoptosis.
Role of p53 in Disease The tumore supression is reduced when the p53 is reduced. p53 also damaged in cells by mutagens (viruses, chemicals, or rediation). Also, p53 in itself can inhibit normal p53 (Blagosklonny, 2002).
p53 Target Gene Recognition
[edit | edit source]p53 regulates the gene transcription binding to double-stranded DNA sequences of its choices. In other words, the p53 protein acts as a transcription factor. Each half-site that the p53 can be described as an inverted pentameric quarter site. The human genome is found to have a high probability binding loci that contains half sites without insertion. Within p53C domains, there are 4 spots that bind to response elements which increase upon binding to full length proteins. The teramers also demonstrate different areas of binding which is formed through a symmetrical dimer. The interface is stabilized within the loops that bind by hydrophobic and water mediated polar contacts. Weak protein-protein interactions are observed within the dimers that can reflect to the response elements within the different spacers. The key residues in the DNA interface make direct contact with DNA half sites inside the DNA minor groove. When making contact, it makes salt bridges that help bind. Although the specific contacts with DNA is important, the residue in the DNA domain are disordered sometimes. [4]
p53 Cancer & Mutations
[edit | edit source]Although the p53 may help in repressing tumor, some mutations of the TP53 gene is found in some human cancers. These cancers may have over 17,000 cases of somatic p53 missense mutations. Some causes may be the common ancestor found between the p53 genes. There are different mutant classes amongst the mutants. Contact mutations remove DNA-contact residues, while structural mutations affect residues that are essential in the big picture of the DNA binding surface. The equilibrium unfolding and denaturation allow response elements to find even more mutant classes. B sandwich mutations are found in 1/3 of cancer cases in which the B sandwich cannot collapse with the surrounding structure. The mutation destabilizes the protein losing the hydrophobic interactions. [4] Most of the mutations in p53 are missense mutations that affect the DNA binding domain region of the protein. When this happens, the molecule is not longer able to bind to DNA which results in a loss of function.[5] In other cases it has been found that the molecule still tetramerizes and is still capable of binding to DNA, but it no longer can function as a transcription factor. [1]
In the case of mutations that result in the protein losing its ability to bind to DNA, there is a subgroup of mutations that actually result in a "gain of function" and p53 now has an oncogenic role[5]. It this case, it is believed that p53 binds to the DNA binding domain of p73, thus inactivating p73 by preventing it from binding to its targets. As a result, this inhibits that pathways p73 is responsible for, transactivation and apoptosis, thus further promoting cancer[5] This inactivation of p73 by an alread inactive p53 further results in a decreased response to chemotherapy[5]. P53 inhibits tumor formation through the use of Cell Cycle Arrest, Apoptosis and Senecence. This gene has the ability to induce cell apoptosis as part of its role as a tumor suppressor. P53 becomes activated by damage in the cell and is suppressed by MDM2 and when P53 becomes too activated to be suppressed it is possible for it to activate the apoptosis pathway in the cell, among other pathways. Some of the proteins that activate p53 are the MAPK proteins, ATR, TP53RK, and p14ARF. The acetylation sites on p53 play a key role in controlling the specific p53 activation sites and if both are mutated the mutation causes the loss of ability for p53 to induce cell cycle arrest and apoptosis.
Regulation of p53
[edit | edit source]MDM-2 (murine double minute 2) is the negative regulator of p53 and acts by binding to the first 42 N-terminal residues that contain a transactivation domain.[6] When MDM-2 binds to p53, is causes a α-helix conformation to form which is successful in blocking the transcription and ubiquitinating lysine resides in the C-terminal domain region. When stress signals or abnormal cell division is sensed, the ARF gene is transcribed and its purpose is to bind to MDM-2 and inhibit its activity and allowing p53 levels to rise.[6]
3D Structure
[edit | edit source]Determining the 3D structure of p53 has been very difficult due to the very complicated folding, especially in the C- and N- termini. The best way to study the three dimensional structure of this interesting protein is to look at the structure of each of its domains separately. The domains structure that have been solved so far are the tetramerization domain; DNA binding domain in complex with DNA, a complex with a transactivation domain; and some parts of the regulatory domain in complex with proteins.[4]
References
[edit | edit source]- ↑ a b c d e f g h i j Viadiu, Hector. "Gene Recognition by the P53 Protein Family." CHEM 114A Lecture. University of California, San Diego, La Jolla. 14 Nov. 2012. Lecture.
- ↑ Andrea Bisso1, Licio Collavin1, and Giannino Del "p73 as a Pharmaceutical Target for Cancer Therapy" Laboratorio Nazionale CIB, Area Science Park, Padriciano 99, 34149 Trieste, Italy. 2Dipartimento di Scienze della Vita, Universitàdi Trieste, Via L. Giorgieri 1, 34100, Trieste, Italy/>
- ↑ Levine, Arnold J., and Moshe Oren. "The First 30 Years of P53: Growing Ever More Complex." Nature Reviews Cancer 9.10 (2009): 749-58. Print. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2771725/>
- ↑ a b c d e f g Joerger AC, Fersht AR. The tumor suppressor p53: From structures to drug discovery. Cold Spring Harb Perspect Biol. 2010;2:a000919.
- ↑ a b c d Irwin, Meredith S. "Family Fued in Chemosensitivity." Cell Cycle 3.3 (2004): 319-23. PubMed.gov. Web. 14 Nov. 2012. <http://www.landesbioscience.com/journals/cc/irwinCC3-3.pdf>.
- ↑ a b Belyi, Vladimir A., Prashanth Ak, Haijian Wang, Wenwei Hu, Anna Puzio-Kuter, and Arnold J. Levine. "The Originals and Evolution of the P53 Family of Genes." Cold Spring Harbor Perspectives in Biology, n.d. Web.
