GENtle/Print version
| This is the print version of GENtle You won't see this message or any elements not part of the book's content when you print or preview this page. |
GENtle is an open source software for molecular biology. It is available at http://gentle.magnusmanske.de .
About
[edit | edit source]Rules
[edit | edit source]- Keys are marked like this
- Menus, Buttons etc. are marked like this
- "Double click" always refers to the left mouse button
- "Context menu" always refers to the popup menu that appears on clicking with the right mouse button
Copyright
[edit | edit source]- GENtle is © 2004-2008 by Magnus Manske, licensed under GPL
- This manual is © 2004-2008 by its authors, licensed under GFDL
Setup
[edit | edit source]Installation
[edit | edit source]On Windows, run the file GENtleSetup.exe. It will set up GENtle in a directory of your choice, and also prepare for a clean deinstallation.
Databases
[edit | edit source]There are two types of databases supported by GENtle: File-based (sqlite) and MySQL. By default, a file-based database local.db is set up in the installation directory of GENtle. You can (and should, if access limitations prevent you from working with local.db) create new databases and share them with other users of GENtle in your work group (if you want to).
Database management can be found through the menu "Tools/Manage database", under the tab "Databases".
- A quirk:
- 1. Be careful of using the local db for any real development. I strongly advise that you never put anything into local. That db is in the Gentle program's directory and if, say, you need to reinstall GENtle, or you get a new laptop, you may clobber it. And if someone else takes over after that, you can lose it also. I have seen other strange behavior where local.db gets lost, but can't tie it down for a bug report.
- 2. GENtle copies blank.db and local.db in [your-username]/AppData/Local/VirtualStore/Program Files (x86)/GENtle It then maintains them. The live local.db is the one in AppData/Local/VirtualStore/Program Files (x86)/GENtle. So, you can delete the one in C:/Program Files (x86)/GENtle and you won't lose it. (This is probably why Magnus did it this way. He was trying to prevent data loss.) But, if you switch computers, this one can get lost.
- So the first thing you should do for serious work is to make a new db of your own and set it as your default db. And monitor local regularly for stuff accidentally getting into local. The symptoms of a problem are that a project can't find parts of a project.
File-based databases
[edit | edit source]You can add an existing or create a new database via the appropriate buttons. Select the database name and location in the following file dialog.
MySQL databases
[edit | edit source]These work similar to the file databases, but require more parameters. Also, a MySQL server has to be installed prior to MySQL database creation. Contact your local system administrator about this.
Creating a new MySQL database might require more MySQL privileges than actually using the created database. Thus, after creation, one might want to remove the database entry via the "Remove" button (the database will continue to exist), and add it again with different MySQL privileges.
DNA
[edit | edit source]
Within the DNA module, DNA sequences can be viewed, edited and annotated. It is the central module of GENtle. Two major components of the DNA module are the DNA map and the Sequence map; see there for details.
Open and display DNA
[edit | edit source]A DNA sequence can be opened in one of the following ways:
- Open from a database (see Databases)
- Import from file (see Import)
- Manual input (see Enter sequence)
- Create from another DNA module
Toolbar
[edit | edit source]Several functions and display options can be invoked in the tool bar:
- Enter sequence
- Open sequence
- Save sequence
- Undo
- Cut
- Copy
- Paste
- Toggle linear/circular
- Show/hide open reading frames
- Show/hide features
- Show/hide restriction sites
- Expand (=show only) map
- Toggle edit mode
- Zoom
Detail tree
[edit | edit source]The detail tree, left of the DNA map, shows all parts of the current sequence, including features and restriction enzymes, in a structured fashion. Features and restriction enzymes can be toggled in visibility by a double click, or further manipulated through the context menu.
Special menus
[edit | edit source]| View/Show 3'->5' | Show the complementary DNA strand in the sequence map |
| Edit/Edit ORFs | Change the settings for open reading frame display |
| Edit/Show possible sequencing primers | Opens the Sequencing Primers dialog, which can add possible sequencing primers as features |
| Edit/Remove sequencing primers | Removes all sequencing primers generated by the above function from the sequence |
| Edit/Auto-annotate sequence | Finds features from common vectors and other databases in the current sequence |
| File/Print map | Prints the DNA map |
| File/Print sequence | Prints the Sequence map |
| File/Print report | Prints a brief overview. See Printing |
DNA map
[edit | edit source]
The DNA map is shown for DNA sequences (though a variant is also used in protein module for the schematics display). It shows the linear or circular (e.g., plasmid) DNA sequence as a map.
Display
[edit | edit source]Displayed are
- features (including optional sequencing primers)
- restriction sites
- sticky ends (if any)
- open reading frames (optional)
- sequence name and length (optional, see Options)
- methylation (optional, see Options)
- GC contents (optional, see Options)
Mouse actions
[edit | edit source]| Action on | Mouse button | Function |
|---|---|---|
| Background | left | Mark sequence |
| left (double click) | Open Sequence editor | |
| middle | Show marked DNA in sequence; show current position in sequence if nothing is marked | |
| Feature | left | Move feature display |
| left (double click) | Edit feature (see Sequence editor) | |
| middle | Mark DNA that matches feature | |
| middle (shift pressed) | Extend currently marked area to include the DNA of the feature | |
| Restriction site | left | Move site display |
| left (double click) | Edit enzyme list (see Sequence editor) | |
| middle | Open Restriction Assistant | |
| Open reading frame | left | Mark ORF sequence |
| left (double click) | Mark and show ORF sequence | |
| All | right | Context menu |
Context menu
[edit | edit source]The context menu opens on a click with the right mouse button when somewhere inside the DNA map. The contents of the menu depends on what object in the map you clicked on. Also, depending on the properties of the object, some functions might not be available, for example, amino acids of a feature with no reading frame.
Background
[edit | edit source]| Edit sequence | Opens the Sequence editor |
| Transform sequence | Make sequence inverted and/or complementary |
| Limit enzymes | Limits enzymes to those that cut no more than n times |
| PCR/PCR | Starts the PCR modul |
| PCR/Forward | Starts the PCR modul and generates a 5'->3'-primer |
| PCR/Backward | Starts the PCR modul and generates a 5'->3'-primer |
| PCR/Both | Starts the PCR modul and generates both primers |
| PCR/Mutation | Starts the PCR modul and generates overlapping mutagenesis primers |
| Selection/Cut | Removes the selected part of the sequence and puts it into the clipboard |
| Selection/Copy | Copys the selected part of the sequence into the clipboard |
| Selection/Copy to new sequence | Generate a new DNA sequence entry based on the selection |
| Selection/Show enzymes that cut here | Opens a variant of the Silent Mutagenesis dialog for the selected part of the sequence |
| Selection/Selection as new feature | Generates a new feature for the selected part of the sequence |
| Selection/Extract amino acids | Extracts the amino acid sequence of the selected part of the DNA sequence |
| Selection/BLAST amino acids | Runs a BLAST search for the amino acid sequence of the selected part of the DNA sequence |
| Selection/BLAST DNA | Runs a BLAST search for the selected part of the DNA sequence |
| Sequence map/Save as image | Saves the DNA map as an image file |
| Sequence map/Copy image to clipboard | Copies the DNA map as a bitmap or WMF (see Options) to the clipboard |
| Sequence map/Print map | Prints the DNA map |
| Show/hide ORFs | Toggles the open reading frame display |
| Edit ORFs | Adjusts the open reading frame display |
Restriction sites
[edit | edit source]| Edit restriction enzyme | Add/remove/manage restriction enzyme via the Sequence editor |
| Show/hide enzyme | Toggle visibility for the enzyme (this will affect all restriction sites for that enzyme in this sequence) |
| Remove enzyme | Remove the enzyme from the current selection (this will affect all restriction sites for that enzyme in this sequence). This will not work for automatically added enzymes (see Options) |
| Mark restriction site | Marks the recognition sequence of that enzyme at that restriction site |
| Mark and show restriction site | Marks the recognition sequence of that enzyme at that restriction site and shows it in the sequence |
| Online enzyme information | Opens the ReBase page for that enzyme |
| Add to cocktail | This adds the enzyme to the restriction cocktail (see Restriction Assistant) |
| Add to cocktail | This adds the enzyme to the restriction cocktail (see Restriction Assistant) and starts the restriction |
Features
[edit | edit source]| Edit feature | Edit the feature properties (see GENtle/Sequence editor) |
| Hide feature | Hide the feature from display |
| Delete feature | Delete the feature |
| DNA Sequence/Mark feature sequence | Mark the DNA sequence that matches the feature |
| DNA Sequence/Mark and show feature sequence | Mark the DNA sequence that matches the feature and shows it in the sequence |
| DNA Sequence/Copy (coding) DNA sequence | Copies the DNA sequence that matches the feature to the clipboard |
| DNA Sequence/This feature as new sequence | Generates a new DNA sequence based on the feature |
| DNA Sequence/BLAST DNA | Runs a BLAST search for the DNA of the feature |
| Amino acid sequence/Copy amino acid sequence | Copies the amino acid sequence of the feature to the clipboard |
| Amino acid sequence/As new entry | Generates a new protein entry based on the amino acid sequence of the feature |
| Amino acid sequence/Blast amino acids | Runs a BLAST search for the amino acid sequence of the feature |
Open reading frames
[edit | edit source]| As new feature | Generate a new feature from the ORF, with the appropriate reading frame and direction |
| DNA sequence/Copy DNA sequence | Copies the DNA sequence of the ORF to the clipboard |
| DNA sequence/As new DNA | Generates a new DNA sequence entry based on the DNA sequence of the ORF |
| DNA sequence/BLAST DNA | Runs a BLAST search for the DNA sequence of the ORF |
| Amino acid sequence/Copy amino acid sequence | Copies the amino acid sequence of the ORF to the clipboard |
| Amino acid sequence/As new AA | Generates a new protein entry based on the amino acid sequence of the ORF |
| Amino acid sequence/BLAST amino acids | Runs a BLAST search for the amino acid sequence of the ORF |
Sequence map
[edit | edit source]
The sequence map is used by most GENtle modules. It shows sequences of DNA or amino acids, as well as primers, features, restriction sites and more. The basic behaviour, however, is always similar.
Clicks
[edit | edit source]A double click usuaqlly opens the editor for the sequence.
Context menu
[edit | edit source]The available functions in the context menu vary with the module the sequence map is used in, its state, and selection.
| Edit sequence | Turn on edit mode |
| Transform sequence | Invert and/or complement the sequence (DNA module only) |
| Limit enzymes | Limit enzymes so that only enzymes below a certain number of cuts in the sequence is shown (DNA module only) |
| PCR | Compare DNA map |
| Selection | Compare DNA map |
| Copy as image | Copies the sequence map as a bitmap to the clipboard (Caveat : Such a bitmap can take up a huge amount of memory, depending on the length of the sequence) |
| Save as image | Saves the sequence map in one of several image formats |
| Print sequence | Prints the sequence |
Keys
[edit | edit source]The whole sequence can be marked by Ctrl-A. The Find dialog can be invoked by Ctrl-F. Both functions can also be called upon through a menu.
In the DNA and PCR modules, the amino acid reading frame can be toggled by keys like this:
- Ctrl-1 = reading frame 1
- Ctrl-2 = reading frame 2
- Ctrl-3 = reading frame 3
- Ctrl-4 = reading frame 1, complementary strand
- Ctrl-5 = reading frame 2, complementary strand
- Ctrl-6 = reading frame 3, complementary strand
- Ctrl-7 = all reading frames, one-letter code
- Ctrl-8 = known reading frames only (from the features)
- Ctrl-0 = hide amino acids
- Ctrl-W = three-letter code (not when displaying all reading frames)
- Ctrl-Q = one-letter code
Edit mode
[edit | edit source]Display and edit mode can be toggled by F2 or the toolbar. During editing, the sequence display is maximized, and the DNA map is hidden, improving ease of edit. Depending on the current module, only some keys are allowed (in the DNA module, "A", "C", "G", and "T") by default; any other key will trigger a request to allow all keys for that sequence, for that session. The cursor can be moved similar to that in a text editor. Insert and overwrite mode can be toggled, except for some modules like PCR or Sequencing, where overwrite mode is mandatory. In these modules, backspace and delete are diasbeled as well.
When editing a primer in PCR mode, the "." key copies the base at the current position from the 3'→5' or 5'→3' sequence, respectively.
Horizontal mode
[edit | edit source]In some modules, the sequence display can be toggled to horizontal. This can enhance visibility. Printing, however, is always done in standard ("vertical") mode.
Protein
[edit | edit source]
In this module, amino acid sequences (peptides/proteins) can be viewed, edited and annotated. It uses a sequence map as main display, and a multi-purpose overview display at the top.
Toolbar
[edit | edit source]Several functions and display options can be invoked in the tool bar:
- Enter sequence
- Open sequence
- Save sequence
- Print sequence
- Undo
- Cut
- Copy
- Paste
- Plot (shows a plot within the sequence map)
- Horizontal mode
Function display
[edit | edit source]The smaller display on the top can show several types of information:
| Data | Shows some basic data that has been calculated from the sequence |
| Description | Shows the sequence description |
| Scheme | Shows a DNA map-like layout of the whole protein |
| AA weight | Shows a plot of the molecular weight of the individual amino acids |
| AA isoelectric point | Shows a plot of the isoelectric point of the individual amino acids |
| Hydrophobicity | Shows a plot of the local hydrophobicity of the amino acids nearby |
| Chou-Fasman plot | Shows a detailed Chou-Fasman-plot |
Special menus
[edit | edit source]| Edit/Photometer analysis | Invokes the respective calculator |
| Edit/'Backtranslate' DNA | Attempts to generate the DNA sequence which codes for this amino acid sequence, using the full range of IUPAC base letters |
| Edit/Proteolysis assistant | Invokes the proteolysis assistant |
PCR and Primer Design
[edit | edit source]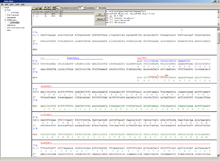
This module allows for designing primers and running virtual PCRs. It can be started from a DNA module via context menu of the DNA or [../Sequence map#Context menu|sequence]] map, or through Tools/PCR. If a sequence is selected in the DNA module, one or more primers can be generated automatically upon startup of the PCR module. These will only be rough suggestions, and are in no way optimized by default.
Toolbar
[edit | edit source]- Enter new primer
- Open primer/sequence
- Print PCR
- Add a primer (you will have to open or enter the primer first)
- Export a primer (generate its sequence)
- Edit mode
- Show/hide features
- Polymerase running length
- Horizontal mode
The polymerase running length is the number of nucleotides the polymerase is allowed to run during the PCR in the elongation step. This is usually measured in minutes, but each polymerase runs at a different speed, which is why this information is given here in nucleotides. The value is initially computed automatically, but can be changed manually.
Primer list
[edit | edit source]The primer list (the upper left) shows all primers used in this PCR, as well as certain key properties of these. Selecting one of these primers will show more detailed information in the box on the right (see here for details). Double-clicking one of the primers will mark and show that primer in the sequence. A selected primer can be removed through the Remove button, or edited via the Edit button. A selected primer can also be exported via the Export button in the toolbar; a new sequence will be generated for that primer.
- Caveat : The generated sequence is not stored anywhere automatically, it needs to be saved manually!
- Caveat : To add a primer, use the Add button in the toolbar, or the Selection as new primer context menu. Merely editing the sequence (see below) is for editing existing primers only, it will not create new ones!
Sequence
[edit | edit source]The sequence consists of the following lines:
- Features of the template DNA (can be turned off in the toolbar)
- 5' primer
- Template DNA (5'→3')
- Amino acid sequence of the template
- Template DNA (3'→5')
- 3' primer
- Restriction sites of the resulting DNA
- Resulting DNA (shown in green)
- Amino acid sequence of the resulting DNA
Some special functions and properties of the PCR sequence display:
- The amino acid reading frame can be set as described here. This will affect both amino acid sequences shown (template and result).
- Only the two primer sequences can be edited; overwrite mode is mandatory, and deleting is disabled.
- To delete a nucleotide, overwrite it with Space.
- The "." key will copy the matching template nucleotide to that position in the primer sequence.
- Matching primer nucleotides (that is, matching with the template) are shown in blue, mismatches in red.
- If (when not in edit mode) an empty span of the primer sequence is selected, it can be turned into a new primer via the context menu (Selection as new primer).
- The sequence of a restriction site can be inserted left or right of a selection (in edit mode, right or left of the cursor) via the context menu. A selection dialog for the desired enzyme will appear.
- A silent mutation can be introduced via the context menu.
Finally, the resulting DNA or amino acid sequence (the green sequence, which will be the one generated by the PCR) can be copied to the clipboard or generated as a new sequence (containing all features, restriction enzymes etc.) via the context menu.
Sequencing
[edit | edit source]
The sequencing module allows to view the data recorded by a sequence analyser. The data is loaded by importing the appropriate .abi/.ab1 file.
Display
[edit | edit source]The data is displayed in the main sequence window. The text window on the upper right shows data stored in the file. On the left side, the following display options for the sequence are available:
| Help lines | Gray vertial lines down from each sequence letter to the baseline. These can help to identify which letter belongs to which peak |
| Invert&complement | Shows the sequencing complement/inverted. Useful for Alignments |
| Scale height | Sets the height of the graphic display [unit in text lines] |
| Scale width | Sets the graphical points per data value. Default is 2; 1 would mean one pixel width per data point |
| Zoom | Sets the zoom factor for the data; useful to see small peaks |
Toolbar
[edit | edit source]- Enter new sequence
- Open sequence
- Save sequence (see caveats)
- Copy sequence to clipboard
- Horizontal mode
Caveats
[edit | edit source]- Editing works in overwrite-mode only
- Saving will only store the sequence in the database, not the sequencer data (the peaks), due to memory concerns.
Alignments
[edit | edit source]
The alignment module displays alignments of DNA and amino acid sequences. It can be invoked through Tools/Alignment or Ctrl-G.
Settings dialog
[edit | edit source]The settings dialog will be invoked upon starting the module, or through the "settings" button in the toolbar. The sequences to align, their order, and the alignment algorithm and its parameters can be chosen here. The following algorithms are available:
| Clustal-W | This (default) algorithm generates alignments of high quality, but is rather slow for simple alignments, and sometimes stumbles over local alignments. |
| Smith-Waterman | An internal, fast, but simple algorithm for local alignments, that is, aligning one or multiple short sequences againast a long one. The long sequence has to be the first. It works great for checking sequencing data against the expected sequence. |
| Needlemann-Wunsch | An internal, fast, but simple algorithm for global alignments, that is, aligning sequences of roughly the same length (e.g., different alleles of a gene). As with Smith-Waterman, all alignments are made against the first sequence. |
Caveat : Clicking OK in this dialog will recalculate the alignment; the previous alignment and all manual changes made to it will be lost.
Toolbar
[edit | edit source]Several functions and display options can be invoked in the tool bar:
- Enter sequence
- Open sequence
- Save sequence
- Print sequence
- Settings
- Horizontal mode
- Middle mouse button function
View menu
[edit | edit source]Some display options can be combined with each other:
- Bold (shows characters in bold)
- Mono (black-and-white mode)
- Conserved (shows characters that match the one in the first line as dots)
- Identity (toggles the "identity" line)
Some of them exclude one another:
- Normal (shows colored text on white background)
- Inverted (shows white text on colored background)
Some other display options are planned, but not implemented as of now.
Sequence display
[edit | edit source]The sequence map can be altered through the context menu. These changes will only alter the display, not recalculate the alignment.
- Lines can be moved up or down
- Features for each line can be shown or hidden. By default, features for the first line are shown, features of the other lines are hidden.
- Gaps can be inserted or deleted, in this line, or all except this line. One of these four possible functions is additionally assigned to the middle mouse button; this setting can be changed in the toolbar.
- A double click on a character (not on a gap) opens the "source" window for that sequence (if available), marks and shows the position that was clicked in the alignment. This can be helpful for checking a sequencing.
- Sequences can be marked across multiple lines, then formatted via the Appearance context menu.
Sequences can not be edited within the alignment module. For that, you will have to edit the original sequence, then re-run the alignment.
Notes
[edit | edit source]- Legend for the ClustalW consensus line:
- * = identical or conserved residues in all sequences in the alignment
- : = indicates conserved substitutions
- . = indicates semi-conserved substitutions
Calculators
[edit | edit source]
The calculator module can be invoked via Tools/Calculator. It contains several specialized spreadsheet-based calculators for typical tasks in molecular miology. The editable fields are shown in blue, the (major) results of the calculation are shown in red.
Ligation calculator
[edit | edit source]This calculator gives the amount (in µl) of vector and insert for a ligation, based on the length and concentration of each respectively, their desired ratio and total mass of DNA. A typical ratio of insert:vector is 3:1
DNA concentration calculator
[edit | edit source]This calculator gives the amount and purity of DNA based on photometric absorption at 260 and 280 nm, respectively, as well as the dilution (in case one measures a 1:100 dilution of the original DNA sample) and a correction factor for different types of nucleic acids.
Protein calculator
[edit | edit source]This calculator gives the amount and purity of peptides/proteins based on photometric absorption at 250 and 280 nm, respectively, as well as the molecular weight of the peptide, the layer thickness of the cuvette used, and the number of tryptophanes, tyrosines and cysteines in the peptide.
Data
[edit | edit source]This shows a codon table and a reverse codon table, both for standard code. This page can not be edited.
Virtual Gel
[edit | edit source]
A virtual agarose (DNA) gel can be generated or expanded via the Restriction Assistant. A new virtual gel will be created if none exists; otherwise, the existing one will be expanded.
Within the gel viewer, gel concentration can be varied. Also, labeling can be turned on/off. Gels can be printed, or saved/copied as an image.
The name of a lane can be changed by double-clicking on the lane.
You can chose one of several DNA markers. If your favorite marker is not there, you can file a feature request or add your own directly.
Image Viewer
[edit | edit source]
The Image Viewer module can be invoked via Tools/Image viewer. It can display images, such as gel photos, print them, or save them in another image format.
The viewer can read and write common formats, such as JPG, TIF, BMP, GIF, etc. In addition, it can read the IMG format used by the BioRad Molecular Analyst software.
The directory can be selected via the upper left button. The files in that directory are shown below. A single click on a file displays the image.
The context menu of the image contains entries to save or print the image, or copy it to the clipboard. For saving, PNG, TIF, BMP, and JPG are available formats, with PNG being the default, as it has the best lossless compression.
Labels of IMG images are shown on screen, print, and saved images by default. This can be changed through the "Show labels" checkbox beneath the file list.
An image can be inverted (black <=> white) through the "Invert" checkbox.
Web interface
[edit | edit source]The GENtle web interface lets you access DNA and amino acid sequences from NCBI, as well as publications listed at PubMed. The interface also covers BLAST searches.
NCBI
[edit | edit source]Chosing Nucleotide or Protein, entering a sequence name/keywords, and hitting Search/ENTER will show the NCBI search results for that query. More results (if any) can be browsed with >>.
Double-clicking an entry will download and open the (annotated) sequence.
PubMed
[edit | edit source]The PubMed option gives new entry fields for author(s) (written "Lastname Initials", separated by ","), and date limitations (years), as well as a result sort option.
Double-clicking an entry will open a web browser window with the respective PubMed abstract page.
BLAST
[edit | edit source]Running a BLAST search for a DNA or amino acid sequence will open a new tab in the web interface, showing a countdown for the time the BLAST results are expected to arrive. Once loaded, the results are displayed as simple alignments.
Double-clicking an entry will open the found sequence.
Ligation
[edit | edit source]
The ligation dialog is a means for virtually ligating two (or more) DNA fragments. It can be invoked via Tools/Ligation or Ctrl-L.
The left list shows all potential DNA sequences to be ligated. Some of these are automatically selected, but selection can be manually changed. The right list shows the possible products of a ligation of the selected sequences. Some circular products will be shown in two forms (A-B and B-A), which only differ visually.
The selected products will be generated as new sequences on clicking the Ligate button.
Options
[edit | edit source]Global program options can be altered via Tools/Options.
Global settings
[edit | edit source]
| Option | Description |
|---|---|
| Language | Currently English and German are available |
| Enhanced display | Can be turned off on machines with very show graphics |
| Show sequence title | Displays the sequence title in the DNA map |
| Show sequence length | Displays the sequence length in the DNA map |
| Load last project on startup | Automatically loads the last used project when starting GENtle |
| Use metafile format | Generates a WMF when copying the DNA map instead of a bitmap |
| Show splashscreen | Shows the GENtle splash screen when starting |
| Check for new version on startup | Checks (and downloads) a new GENtle version via internet on startup |
| Use internal help | Help should open in a browser window by default. If that doesn't work, check this option |
Enzyme settings
[edit | edit source]
Here the gloal enzyme options can be selected. These can be overridden for an individual sequence in the sequence editor, where there is a tab identical to this one.
| Option | Description |
|---|---|
| Use global enzyme settings | Turn most of the other options on this tab on or off globally |
| Join enzymes | In a DNA map, cuts of isoenzymes can be grouped together instead of displayed individually |
| Use color coding | Restriction enzymes can be shown in a color matching their number of cuts in a given sequence. The three buttons to the right of this option each hold a color choice dialog for single, double, and triple cutters. |
| Use min/max cutoff | Shows only enzymes that cut a minimum/maximum times |
| Sequence length | Shows only enzymes with recognition sequences of the selected lengths |
| Use enzyme group | Uses only enzymes from the selected enzyme group |
| Show methylation | Shows DAM and/or DCM methylation in map and sequence, in red |
| Show GC contents | shows the GC contents in the map |
Databases (Open/save/manage)
[edit | edit source]
The GENtle database management dialog is where sequences are stored and retrieved. DNA and amino acid sequences, primers, alignments, and projects all go to databases, which can be local (for one computer only) or shared (used by the whole work group, institute, etc.).
Management
[edit | edit source]The "Management" tab can be reached through the File menu, the Tools/Manage database menu, the Ctrl-O and Ctrl-S keys ("open" and "save", respectively), or the appropriate buttons in the toolbar. The tab consists of two or three parts:
Filter
[edit | edit source]The filter section allows to filter the database entries so the list(s) below show only the matching entries.
The filter text box limits the shown sequences to those whose name (or description or sequence, depending on the checkboxes) contain that text. Multiple search words are separated by a space (" ") and work as a logical AND. Thus, entering "pgex igf" in the filter text box shows only those sequences whose name (or description) contain both the word "pgex" and "igf". The search in not case-sensitive, so searching for "igf" or "IGF" will make no difference.
The checkboxes on the right limit the display to any combination of DNA, protein (amino acid sequences), and primers. If non of these is selected, all types of entries are shown, including alignments. As already described, search for text can be extended beyond the sequence name to description and the sequence itself through two other checkboxes, where description search is enabled as default.
Lists
[edit | edit source]One or two lists are shown, depending on the appropriate checkbox above the left list. The database(s) to search/display can be selected via the drop-down box(es). One list with full width is good for an overview of a single database, whereas two lists are needed for moving and copying entries between databases; also, a search will be run on both databases simultaneously.
Entries will be sorted alphabetically. Every entry has a small icon associated with its type. There are icons for DNA, amino acid sequences, primers, and alignments. There is also a project icon, but these will only be shown when opening/saving a project.
A single entry can be selected by clicking with the left mouse button. When opening a file, a double click or pressing RETURN on a selected entry will open it. Multiple entries can be selected by dragging a rectangle with the mouse, or by holding down the SHIFT and/or CTRL keys. A multiple selection can be opened via RETURN.
Grabbing selected entries with the left mouse button and dragging them into the other list will move these entries to that database. To copy these entries, hold down the CTRL key when releasing the left mouse button over the target list.
Selected entries can be opened, renamed, and deleted via their context menu.
Save
[edit | edit source]If you save an entry to a database, there will be an additional line below the lists. It consists of a drop-down box with the database to save the entry to, and a text box for the name. The name of the database is remembered if you originally opened that entry from a database, otherwise the standard database is the default.
Saving an entry to a database where an entry with that name already exists will lead to the following:
- If the sequence of the entry in the database is exactly the same as the sequence of the entry you're trying to save, a message box will ask you if you really want to overwrite that entry.
- If the sequence of the entry in the database differs from the sequence of the entry you're trying to save, a message box will tell you that this action was prevented. This will avoid accidental overwriting of an entry with a different sequence. If you are very certain you want to replace that entry, you will have to delete the entry in the database manually via the context menu, as described above.
Databases
[edit | edit source]Currently, GENtle supports sqlite and MySQL databases, both of them freely available. Each has different advantages and disadvantages, though both are integrated seamlessly into GENtle. Once set up, all functions are available on all databases, no matter the type.
The "Databases" tab keeps a list of all the databases that can be accessed. New databases can be created, and existing can be added to or removed from that list. The exception is the local database, which is essential for the functioning of GENtle and therefore can not be removed. Removal of a database will not delete the database itself, only the entry in the list.
One of the databases in the list is the default database. The default database can be set by selecting its entry in the list, then clicking the As Default button. The default database can carry shared enzyme groups.
Sqlite
[edit | edit source]Sqlite is already integrated in GENtle, so no separate installation or setup of any kind is required. A sqlite database consists of a single file with the ending ".db". For each GENtle installation, a database ("local.db") is automatically created. New sqlite databases can be created, or existing ones added to GENtle, on the "Databases" tab in the dialog. To take such a database with you (e.g., for use at home or on a laptop), just copy the ".db" file. While sqlite databases are easy to set up and maintain, sharing them across a network tends to be slow, depending on the size of the database.
MySQL
[edit | edit source]MySQL is a professional client/server database system that will reliably store and serve millions of entries. It is ideal for shared databases, as even a huge number of stored sequences will not slow it down significantly, even across a network. However, there are some steps required to use MySQL databases with GENtle:
- A "server" computer on your network, that is, a computer that is running most of the time, and preferably is not used for direct work. If the server is not running, or disconnected from the network, no one will be able to access the MySQL database and the sequences stored in it!
- The MySQL server software (4.1 works fine, other versions will likely do as well), which available for free here.
- Someone to configure the MySQL server (not as complicated as it sounds)
Once the MySQL setup is complete, MySQL databases can be created (by one) and added to all the GENtle clients that should have access.
Import
[edit | edit source]The import dialog is a standard "file open" dialog. It can be invoked via Files/Import or Ctrl-I.
Multiple files can be chosen to be imported in a row. GENtle will automatically try to determine the file type, but also a file type can be chosen manually.
Supported formats include:
- GenBank
- GenBank XML
- FASTA
- ABI/AB1 (popular sequencer output format)
- PDB (a 3D format, import as annotated sequence)
- Clone (old DOS program, proprietary format)
- Numerous other sequence formats that will be imported as "sequence only", without annnotations, features etc.
- Comma-separated values (CSV) from various machines (HPLC/FPLC/photometer) for graphs and plots
Enter sequence
[edit | edit source]
This dialog to enter a sequence manually can be invoked via File/Enter sequence or Ctrl-N.
Beside the sequence, to be typed or pasted into the large text box, one can enter a title (name) for that sequence, and choose a type.
Types available are:
- DNA
- Amino acid sequence
- GenBank
- (GenBank) XML
- Primer
When chosing DNA, amino acids, or primer, all non-sequence characters, like blanks and numbers, are automatically removed.
- Note : A primer has to be given the type "Primer", otherwise it will be added as DNA.
Find dialog
[edit | edit source]
The Find dialog in DNA and amino acid sequence can be invoked via Ctrl-F or Edit/Search. It displays can find a string in
- the current sequence
- a feature name
- a feature description
In DNA sequence display, it also look in
- the reverse sequence
- the translated amino acid sequence(s)
- restriction enzyme names
The search is commenced automatically after changing the search string, if it is three or more characters long. For shorter search queries, the Search button has to be clicked.
Single-clicking on a search result will select and display the result in the sequence. A double click will exit the dialog, and open the sequence editor for features, or the enzyme management dialog for restriction enzymes.
Sequence editor
[edit | edit source]
The sequence editor holds the key to several properties of a sequence. It consists of several tabs, depending on the type of sequence, which can be DNA or amino acid.
Properties
[edit | edit source]Here, the title and description of the sequence can be altered. As for feature descriptions, the sequence description will make http references clickable.
For DNA sequences, sticky ends can be entered.
Features
[edit | edit source]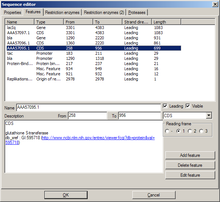 |
 |
This tab shows a list of all features of the sequence. Features can be added, edited, and deleted. Most of the settings should be self-explanatory.
- The setting reading frame is only available when the type is set to "CDS" ("coding sequence").
- A leading sequence is read 5'→3'; leading unchecked, 3'→5'
- Edit feature will invoke an additional "Edit feature" dialog
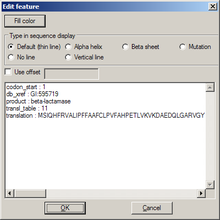
Edit feature
[edit | edit source]- Fill color is the color of the feature; it will invoke a color choice dialog
- Type in sequence display determines how that feature is drawn in the sequence map
- Use offset sets the numbering for the first amino acid of the feature; useful if the feature marks a part of a protein
The list box below contains original data from GenBank format import.
Restriction enzymes
[edit | edit source]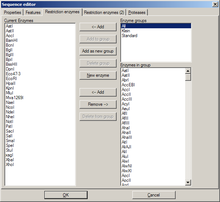 |
 |
When editing a DNA sequence, two tabs with settings for restriction enzymes are available. The first one is identical to the enzyme management dialog. The second one is identical to the global enzyme settings tab, but contains the settings for this sequence alone. By default, its options are disabled, and the global options are used. By activation the options here, global settings are overridden.
Proteases
[edit | edit source]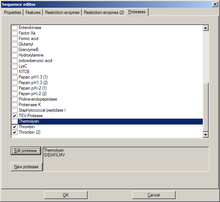
This tab holds a list of available proteases. Potential cleavage sites for selected (checked) proteases are shown in the sequence (not in the DNA map).
New proteases can be added similar to the following examples:
- Example : "Thermolysin"
- Sequence for this protease : "!DE|AFILMV"
- Explanation: "Not D or E", "cut", "then A, F, I, L, M, or V"
- Example: Proline-endopeptidase
- Sequence for this protease : "HKR,P|!P"
- Explanation : "H, K, or R", "then P", "cut", "then not P"
Restriction Assistant
[edit | edit source]
The Restriction Assistant can be invoked via menu Tools/Enzyme Assistant, or through a click with the middle mouse button on a restriction site in the DNA map. For the latter, the selected enzyme is automatically selected in the list of "Available enzymes" (left). This list depends on the selections "Group" and "Subselection". It can be sorted by enzyme name or number of cuts by clicking on the respective column title. For a selected enzyme, the resulting fragments are shown in the lower left list.
The list on the right shows the contents of the "restriction cocktail", the enzymes already selected for cutting. The resulting fragments for these enzymes together are shown in the lower right list. The enzyme selected in the left list can be put in the cocktail via Add to cocktail; all enzymes from the left list can be added at once via Add all. An enzyme can be removed from the cocktail by selecting it in the right list, then via Remove enzyme.
Do not create fragments below ___ base pairs, when selected, limits the fragments generated to a minimum size. Done exits the restriction assistant while preserving the changes mage to the cocktail, whereas Cancel will void all changes made.
Start restriction (the scissors symbol) will initiate the simulated restriction. The result of this can be influenced by several further settings:
- Create fragments will generate the actual DNA sequences with their blunt/sticky ends that will result from a digestion with the cocktail. This option is pre-selected.
- Add to gel will add the fragments to a virtual gel, together in one lane.
- One lane each will alter the above so that each enzyme gets its own lane.
- Partial restriction will add all possible fragments to a virtual gel lane, simulating a partial (incomplete) restriction. The option One lane each is not available when Partial restriction is checked.
The restriction cocktail will be preserved so you can cut another DNA with that very enzyme combination, which is useful for an upcoming Ligation.
Projects
[edit | edit source]A project in GENtle is a collection of sequences that belong together, even if they are in different databases. Projects can be
- loaded via File/Load Project or F11
- saved via File/Save Project or F12
- closed via File/Close Project
Depending on the options, the last used project is automatically opened when GENtle starts.
Projects consists of a list of sequences, not the sequences themselves. If a sequence is renamed, moved or deleted, GENtle will dispay a warning next time a project containing that sequence is opened.
For efficient use of sequencing primers, one can create a project that contains all available sequencing primers, and then refer to that project in the Sequencing Primers dialog.
Edit primer dialog
[edit | edit source]
This dialog assists in optimizing a primer. For that reason, many variants of the primer are generated and can be examined.
The center line of the dialog shows the current variant of the primer; details of that variant are shown in the upper right box. OK will end the dialog, committing that variant to the PCR module. Cancel will end the dialog and not change the PCR module. Reset will return the primer in the dialog to the variant the dialog was originally started with.
The list in the lower half of the dialog contains an automatically generated list of variants of the current primer, sorted by an arbitrary score. The "region" of variants can be influenced by multiple settings in the upper left quarter of the dialog. Available settings include:
- The variation of the 5'-end of the primer to the right and to the left.
- The variation of the 3'-end of the primer to the right and to the left.
- The minimum and maximum length of the primer.
- The minimum and maximum melting temperature of the primer.
Any change of these settings will trigger a recalculation of possible variations. These variations are then evaluated and shown in the list in the lower half of the dialog. Double-clicking one of the variations will change the current variation in the center line, and the properties display in the upper right quarter of the dialog.
Properties display
[edit | edit source]This will display:
- The primer sequence in 5'→3' orientation
- The ΔH and ΔS values
- The length and GC contents of the primer
- The melting temperature, calculated according to the Nearest Neighbour method (usually best results, but only for longer primers)
- The melting temperature, calculated according to the salt-adjusted method (medicore results)
- The melting temperature, calculated according to the GC method (simplicistic)
- The highest self-annealing score (arbitrary) and the display of that annealing
- Caveat : Calculating primer melting temperatures is tricky. If one of the three methods gives a totally different result than the other two, ignore it. Also, the melting temperature is only calculated for the 3'-end of the primer that anneals with the sequence!
Printing
[edit | edit source]Sequence and DNA maps can be printed via the respective context menus or the File menu.
For DNA sequences, a report can be printed via File/Print report. It contains the DNA map and a list of the features annotated in the sequence. This can be useful for a detailed overview of the sequence where the sequence itself is not required.
Enzyme management
[edit | edit source]
The enzyme editor for enzyme management, both globally and per DNA sequence, is divided into three lists:
- A list of enzyme groups (top right)
- A list of enzymes in that group (bottom right)
- A list of current/temporary enzymes (left)
Enzymes can be copied into/removed from the left list through the <--Add and Remove--> buttons. Enzymes can be deleted from a group (except All) via Delete from group, or added via New enzyme. A double click on an enzyme name in either list shows an enzyme properties dialog.
Enzymes from the left list can be added to a new or existing group via the respective buttons. All enzymes from a group can be added to the left list, and a group can be deleted.
You can share enzyme groups with other GENtle users on your intranet via a commonly shared database. Create an enzyme group as "Database name:Enzyme group name", and it will be available to everyone using that database on the next start of GENtle. Note that when creating the enzyme group, use the name of the database as it appears in your local installation (maybe "Shared", "Shared0", "Shared (2)" etc.).
Sequencing Primers
[edit | edit source]
The sequencing primers dialog can add possible sequencing primers as features to a DNA sequence. What primers to add can be specified:
- The minimum alignment (3') of a primer to the sequence. This means exact annealing!
- The database to search for primers. All primers from that database will be considered.
- Alternatively, use all primers that are part of a project in that database. That way, a range of primers across databases can be specified in a project and be considered as sequencing primers here.
- Primers that run in 5'→3' or 3'→5' direction.
You can also have the dialog remove old sequencing primers from the sequence. This can also be done manually through Edit/Remove sequencing primers in the DNA module. Note: Sequencing primers, if not removed, will be saved as features together with the sequence; they can still be removed lated, though.
Sequencing primers will display as yellow features, where the shade of yellow depends on their direction. The actual sequencing primer feature is only as long as the 3' annealing of the primer, so the primer might actually be longer than the feature towards the 5' end. For details, see the feature description, which contains the original primer sequence, among other data.
Silent Mutagenesis
[edit | edit source]
This dialog can find restriction enzymes that cut in a marked DNA sequence (context menu Selection/Show enzymes that cut here in the DNA module). It can also find alternate versions of the DNA which will translate into the same amino acid sequence, but contains a new restriction site (silent mutation). A chosen enzyme/mutation will appear in the sequence (DNA or primer, respectively) upon OK.
The results can be changed by
- changing which enzyme group to search
- limiting the number of times an enzyme may cut in the whole sequence
- limiting the number of mutations needed for a restriction site to manifest (PCR module only)
Automatic annotation
[edit | edit source]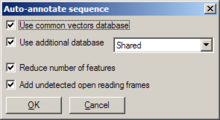
The automatic annotation feature can search a database of standard vectors (included with the GENtle package), and (optionally) a user-generated database, for feature sequences that are found in the currently opened DNA sequence. Recognized features are then annotated in the current sequence.
Invoked through Edit/Auto-annotate sequence or F9, a dialog opens, offering various settings:
- Whether or not to search the common vectors database
- Whether or not to use a user-generated database (and, if so, which one)
- Whether or not to reduce the number of generated features (recommended; otherwise, a lot of features are annotated)
- Whether or not to add unrecognized open reading frames as features
FAQ
[edit | edit source]FAQ - frequently asked questions.
- Q: This manual sucks! And I want one in my language! What can I do?
- A: It's a wiki! You can edit any page and improve the manual yourself. You can even start and maintain a new translation (contact the programmer through this page).
- Q: Why does GENtle try to connect to the internet all the time?
- A: An internet connection is mandatory for BLAST- and ReBase-searches, as well as for the online tools.
- A: At the beginning of each GENtle session, a check for possible updates takes place, which also requires an internet connection. This can be turned off in the Tools/Options menu.
- Q: Why can't I perform a BLAST search for the amino acids coded by the selected DNA sequence?
- Q: Why can't I extract amino acids from the selected DNA sequence?
- A: A reading frame must be selected.
- Q: There's a problem with multiple users using GENtle on my Windows machine.
- A: You might have to allow write access to "local.db" in the GENtle directory for all users involved.
- A: You can turn off the "registry settings bug" via Tools/Options/Register protocols and extensions on startup
- Q: New updates download, but don't install.
- A: You need write access to the GENtle program directory. Alternatively, ask your friendly administrator to download the latest version of GENtle here and install it for you. Remind him that he'll have to do this every few weeks, which might convince him to give you write access...
- Q: I found a bug! And I want new functions as well! Can my favourite protease/marker/restriction enzyme be in the default installation? Where can I donate huge amounts of money for a poor programmer?
- A: Add a bug report or a feature request right here in the wiki, or contact the author of GENtle through the GENtle site.
New proteases to add!
How?
- Open any DNA or amino acid sequence
- Double-click the sequence
- Go to "Proteases"
- Click on "New protease"
- Enter name and sequence (one of the following)
- PreScission
- L,E,V,L,F,Q|G,P
See here for a more detailed description.
